
NEXTFLEX® Rapid DNA-Seq Kit 2.0
Complete library prep solution, including size selection beads
Bundle kits available with library prep reagents and barcodes
Up to 384 barcodes available upon request
- 产品简介
- 工作流程
- 实验数据
- 试剂组分
- 引用文献
- 产品说明书
The NEXTFLEX® rapid DNA-seq 2.0 kit is designed to prepare multiplexed libraries for single or paired-end sequencing using Illumina® platforms. The enhanced NEXTFLEX® kit simplifies workflow by using master mixed reagents and magnetic bead based cleanup, reducing pipetting and eliminating time-consuming steps in library preparation. The NEXTFLEX® rapid DNA-seq kit 2.0 produces library preps with a larger number of unique sequencing reads, allowing for robust genome coverage in challenging GC-rich sequences that can introduce bias.
The NEXTFLEX® rapid DNA-seq 2.0 kit bundle offers all the advantages and reagents of the NEXTFLEX Rapid DNA-seq kit 2.0 and includes NEXTFLEX-HT™ barcodes (8, 48, or 96 reaction size kits) supplied in a single-use format. This allows the lab to tie all the necessary components of the library prep workflow to a single lot for ease of purchasing and tracking of consumables.
产品特点:
Complete library prep solution, including size selection beads
Bundle kits available with library prep reagents and barcodes
Up to 384 barcodes available upon request
Convenient and cost-effective package in a 8, 48, or 96 reaction format
Simplified workflow
Automation compatible with liquid handlers, including the PerkinElmer Sciclone® G3 NGS/NGSx workstations and Zephyr® G3 NGS workstation
Robust genome coverage
Reliable performance with GC-bias and sequencing bias mitigation
Low input requirement for genomic DNA down to 1 ng
For FFPE samples, use the NEXTFLEX® Rapid XP DNA-Seq Kit
Functionally validated with Illumina® sequencing platforms
产品列表:
| 货号 | 产品名称 | 规格 |
| NOVA-5188-01 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | (NO BARCODES) 8 RXNS |
| NOVA-5188-02 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | (NO BARCODES) 48 RXNS |
| NOVA-5188-03 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | (NO BARCODES)96 RXNS |
| NONA-514150 | NEXTflex® Unique Dual Index Barcodes - Set A (Barcodes 1 - 96) | 192 rxns |
| NONA-514151 | NEXTflex® Unique Dual Index Barcodes - Set B (Barcodes 97 -192) | 192 rxns |
| NONA-514152 | NEXTflex® Unique Dual Index Barcodes - Set C(Barcodes 193-288) | 192 rxns |
| NONA-514153 | NEXTflex® Unique Dual Index Barcodes - Set D (Barcodes 289-384) | 192 rxns |
工作流程:
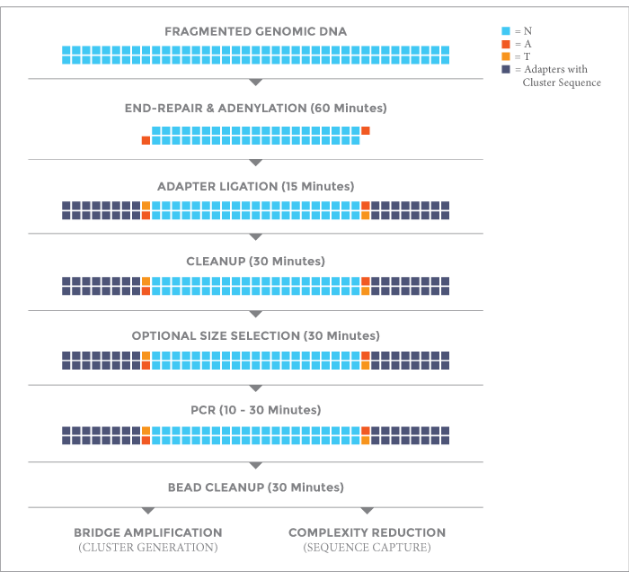
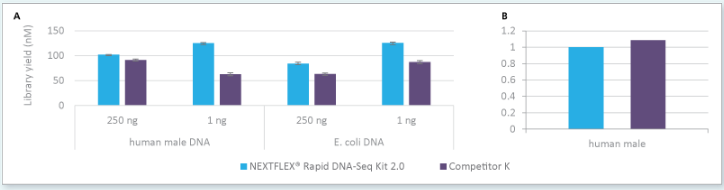
Figure 1: Libraries prepared with NEXTFLEX® rapid DNA-Seq kit 2.0 show higher yield than libraries prepared with Competitor K’s kit using the same number of PCR cycles from a 250 ng and 1 ng input (A) and robust conversion rate (B).
Additional Data between NEXTFLEX® Rapid DNA-Seq 2.0 and Competitor K’s Kit Libraries
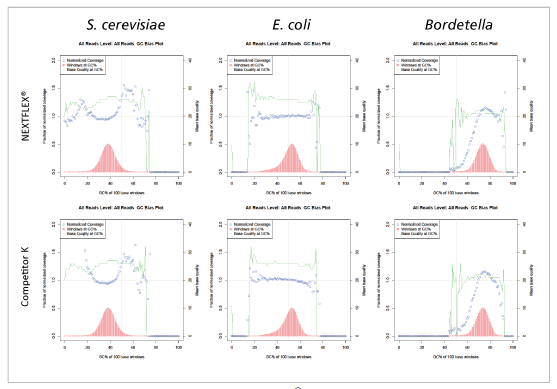
Figure 2: Libraries prepared with NEXTFLEX® Rapid DNA-seq 2.0 shows less GC bias than libraries prepared with Competitor K’s kit.
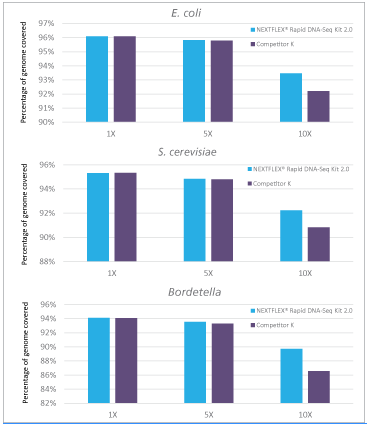
Figure 3: Libraries prepared with NEXTFLEX® Rapid DNA-seq 2.0 shows higher genome coverage than libraries prepared with Competitor K’s kit.
KIT CONTENTS
NEXTFLEX® End Repair & Adenylation Buffer Mix 2.0
NEXTFLEX® End Repair & Adenylation Enzyme Mix 2.0
NEXTFLEX® Ligase Buffer Mix 2.0
NEXTFLEX® Ligase Enzyme 2.0
NEXTFLEX® PCR Master Mix 2.0
NEXTFLEX® Primer Mix 2.0
NEXTFLEX® Cleanup Beads 2.0
Nuclease-free Water
Resuspension Buffer
REQUIRED MATERIALS NOT PROVIDED
1 ng – 1 µg of fragmented genomic DNA in up to 32 µL nuclease-free water.
NEXTflex® barcodes if purchasing a non-bundled kit (includes NEXTflex-HT™ barcodes)
Ethanol 100% (room temperature)
Ethanol 80% (room temperature)
Covaris System (S2, E210) or other method for DNA fragmentation
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) or similar
96 well Library Storage and Pooling Plate (Thermo Fisher Scientific, Cat # AB-0765) or similar
Adhesive PCR Plate Seal (Bio-Rad, Cat # MSB1001)
Magnetic Stand -96 (Thermo Fisher Scientific, Cat # AM10027) or similar
Heat block
Thermocycler
2, 10, 20, 200 and 1000 µL pipettes / multichannel pipettes
Nuclease-free barrier pipette tips
Microcentrifuge
1.5 mL nuclease-free microcentrifuge tubes
Vortex
Shain, A. H., et al. (2015) The Genetic Evolution of Melanoma from Precursor Lesions. The New England Journal of Medicine. doi: 10.1056/NEJMoa1502583.
Campbella, M. A., et al. (2015) Genome expansion via lineage splitting and genome reduction in the cicada endosymbiont Hodgkinia. PNAS. doi: 10.1073/pnas.1421386112.
Gal, C. et. al. (2015) The impact of the HIRA histone chaperone upon global nucleosome architecture. Cell Cycle. 14:1, 123-134. doi:10.4161/15384101.2014.967123.
Yang, Y. A., et al. (2016) FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Research. doi: 10.1093/nar/gkw498.
Palomo, A., et al. (2016) Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. The ISME Journal. doi:10.1038/ismej.2016.63.
Brodie, J., et al. (2016) Characterising the microbiome of Corallina officinalis, a dominant calcified intertidal red alga. FEMS Microbiology Ecology.doi:10.1093/femsec/fiw110.
Givnish, T. J., et al. (2016) Phylogenomics and historical biogeography of the monocot order Liliales: out of Australia and through Antarctica. Cladistics. doi: 10.1111/cla.12153.
Harrisson, K., et al. (2016) Pleistocene divergence across a mountain range and the influence of selection on mitogenome evolution in threatened Australian freshwater cod species. Heredity. doi:10.1038/hdy.2016.8.








