
NEXTFLEX® Methyl Sequencing 1 Kit
Methylome-level assessment with broad genome coverage
Quantify absolute DNA methylation levels
Identify differentially methylated regions (DMRs)
- Product description
- Protocol
- Data
- Kit Contents
- Citations
Methylated DNA Libraries for Illumina® Sequencing
The NEXTFLEX® Methyl-Seq 1 Kit is designed to enrich and prepare single, paired-end and multiplexed methylated DNA libraries for sequencing using Illumina® sequencing platforms. NEXTFLEX® Methyl-Seq utilizes a versatile immune-capture (MeDIP) or methyl-CpG-binding domain (MeCAP) for detection of methylated DNA allowing the user to easily assess the methylation state of the genome.
This kit features Enhanced Adapter Ligation Technology, resulting in library preps with a larger number of unique sequencing reads. This specially designed NEXTFLEX® ligation enzymatic mix allows users to perform ligations with longer adapters and better ligation efficiencies. The NEXTFLEX® Methyl-Seq 1 Kit simplifies workflow by using master mixed reagents and magnetic bead based cleanup, reducing pipetting and eliminating time consuming steps in library preparation.
Flexible Multiplexing Options
NEXTFLEX® DNA Barcodes, NEXTFLEX-96™ DNA Barcodes and NEXTFLEX-HT™ Barcodes can be used for multiplexing libraries prepared with the NEXTFLEX® Methyl-Seq 1 Kit. These kits include adapters containing indexed sequences that offer an improved multiplexing workflow and flexible setup. These barcodes are available in sets of 6, 12, 24, 48, 96 and 384 Illumina-compatible adapters. The ability to pool samples in an efficient way significantly decreases hands on time while providing robust data quality. These barcodes can be used with single, paired-end and multiplex reads.
Features
Methylome-level assessment with broad genome coverage
Make methyl rich libraries using methylated DNA immunoprecipitation (MeDIP) or MBD capture (MeCAP)
Quantify absolute DNA methylation levels
Identify differentially methylated regions (DMRs)
Enhanced adapter ligation technology with NEXTFLEX® Ligation
Flexible barcode options– 6, 12, 24, 48, 96 and 384 unique single-index adapters
Gel-free, bead-based cleanup protocols
Automation-friendly workflow is compatible with liquid handlers
Barcoded adapters for multiplexing contain embedded index sequence
Functionally validated with Illumina® sequencing platforms
Kit Specs
| Cat # | Name | quantity |
| NOVA-5118-02 | NEXTFLEX® Methyl Sequencing 1 Kit | 48 RXNS |
| NOVA-514101 | NEXTflex™ DNA Barcodes - 6 | 48 RXNS |
| NOVA-514102 | NEXTflex™ DNA Barcodes - 12 | 96 RXNS |
| NOVA-514103 | NEXTflex™ DNA Barcodes - 24 | 192 RXNS |
| NOVA-514104 | NEXTflex™ DNA Barcodes - 48 | 384 RXNS |
| NOVA-514105 | NEXTflex™ DNA Barcodes - 96(in 96-well plate) | 768 RXNS |
| NOVA-514106 | NEXTflex™ DNA Barcodes - 96(in tubes) | 768 RXNS |
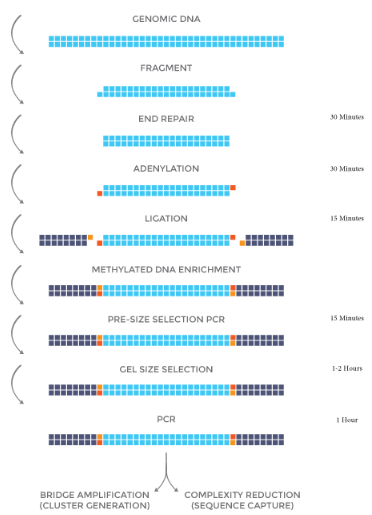
NEXTFLEX® Methyl-Seq 1 Kit Protocol – Option 1
Option 1 is for users who are interested in size selecting a specific range of DNA fragments post ligation with an agarose gel. Proceed to Option 2 for the gel-free protocol.
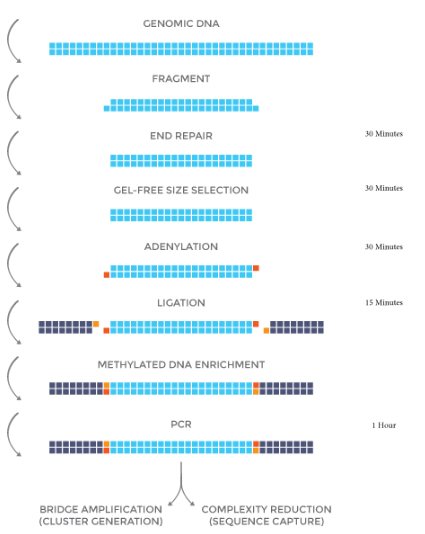
NEXTFLEX® Methyl-Seq 1 Kit Protocol – Option 2
Option 2 is a completely gel-free protocol that utilizes a magnetic bead based cleanup to size select DNA insert fragments between 300 – 400 bp.
Sequence Comparison between Differentiated and Non-differentiated Cells
DNA Methylome, Genome Scale
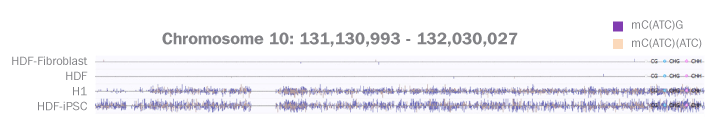
This figure shows that on a genome scale, DNA methylomes of human embryonic stem cells and iPSCs transformed from a human dermal fibroblast cell are similar to one another and highly distinct from the primary somatic cell lines.
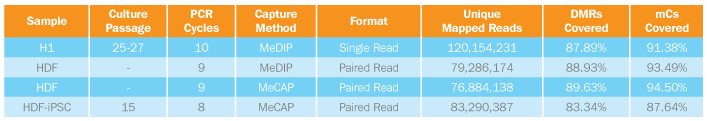
Frequency of DNA Methylation at Both CG and Non-CG sites Suggest iPSCs Resemble ES and are Distinct from Somatic Cells
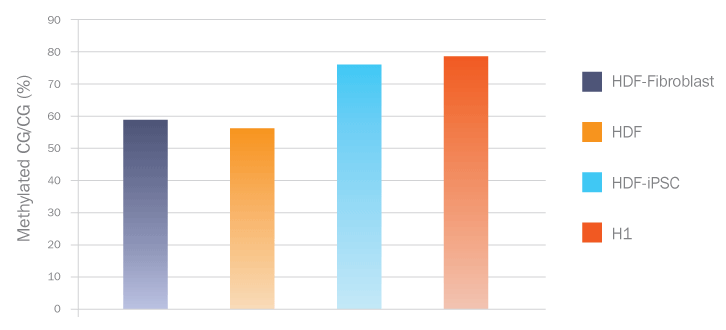
This bar graph shows that high sequence coverage of four DNA methylomes allowed interrogation of 87-94% of genomic methylated cytosines using both MeDIP and MeCAP enrichment. Pluripotent cell coverage is significantly different from somatic cell lines using both enrichment methods.
KIT CONTENTS
NEXTFLEX® Methyl End Repair Buffer Mix
NEXTFLEX® Methyl End Repair Enzyme Mix
NEXTFLEX® Methyl Adenylation Mix
NEXTFLEX® Methyl Ligation Mix
NEXTFLEX® DNA Adapter (25 µM)
NEXTFLEX® Primer Mix (12.5 µM)
NEXTFLEX® Methyl PCR Master Mix
Nuclease-free Water
Resuspension Buffer
REQUIRED MATERIALS NOT PROVIDED
1 µg of fragmented genomic DNA in up to 40 µL nuclease-free water.
NEXTFLEX® DNA Barcodes – 6 / 12 / 24 / 48 (Cat # 514101, 514102, 514103, 514104) or NEXTFLEX-96™ DNA Barcodes (Cat # 514106)
Methylated DNA Enrichment Kit
Methylated DNA IP Kit (Zymo Research Corp., Cat. # D5101) or
MethylMiner™ Methylated DNA Enrichment Kit (Life Technologies, Cat. # ME10025)
Ethanol 100% (room temperature)
Ethanol 80% (room temperature)
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) / or / similar
96 well Library Storage and Pooling Plate (Fisher Scientific, Cat # AB-0765) / or / similar
Adhesive PCR Plate Seal (BioRad, Cat # MSB1001)
Agencourt AMPure XP 5 mL (Beckman Coulter Genomics, Cat # A63880)
Magnetic Stand -96 (Ambion, Cat # AM10027) / or / similar
Heat block
Thermocycler
2, 10, 20, 200 and 1000 L pipettes / multichannel pipettes
Nuclease-free barrier pipette tips
Microcentrifuge
1.5 mL nuclease-free microcentrifuge tubes
Low melt agarose such as Low Gelling Temperature Agarose with a melt point of 65ºC (Boston Bioproducts, Cat # P-730)
1X TAE buffer
Clean razor or scalpel
SYBR Gold (Invitrogen, Cat # S11494)
UV transilluminator or gel documentation instrument
Gel electrophoresis apparatus
Electrophoresis power supply
Vortex
Papers that Cite the Use of the NEXTFLEX® Methyl-Seq Kit
Martínez Pinto, J. (2014) Neonatal exposure to estradiol reprograms the expression of androgen receptor and anti-müllerian hormone [recurso electrónico]: short and long term effects and their relation to the polycystic ovary phenotype. Disponible en http://www.repositorio.uchile.cl/handle/2250/116937.
Winans, B., et al. (2015) Linking the Aryl Hydrocarbon Receptor with Altered DNA Methylation Patterns and Developmentally Induced Aberrant Antiviral CD8+ T Cell Responses. The Journal of Immunology. 194:9, 4446-4457.








