
NEXTFLEX® Bisulfite Library Prep Kit
Compatible with total bisulfite sequencing and reduced representation
Single nucleotide resolution of methylation sites
Uracil insensitive polymerase designed for bisulfite-converted DNA
- Product description
- Protocol
- Data
- Kit Contents
- Citations
Complete Methylome Analysis
The NEXTFLEX® Bisulfite-Seq Kit is a versatile kit designed to facilitate assessment of the methylation state of the genome and simplify workflow by using master mixed reagents and magnetic bead based cleanup to reduce pipetting and eliminate time-consuming steps in library preparation. In addition, the availability of up to 24 methylated NEXTFLEX® Bisulfite-Seq barcoded adapters makes multiplexing simple. This kit features Enhanced Adapter Ligation Technology, resulting in library preps with a larger number of unique sequencing reads. This specially designed NEXTFLEX® ligation enzymatic mix allows users to perform ligations with longer adapters and superior ligation efficiencies.
Focused Sequence Depth
The NEXTFLEX® Bisulfite-Seq Kit with reduced representation utilizes a restriction enzyme that leads to selective amplification of CpG regions, resulting in focused sequence depth. The NEXTFLEX® Msp1 Restriction Enzyme, which is available separately, has been optimized for reduced representation studies using the NEXTFLEX® Bisulfite-Seq Kit. The NEXTFLEX® Bisulfite-Seq Kit can also be used without the MSP 1 Restriction Enzyme for genome-wide methyl-seq analysis, including under-represented CpG regions.
Features
Compatible with total bisulfite sequencing and reduced representation
Single nucleotide resolution of methylation sites
Uracil insensitive polymerase designed for bisulfite-converted DNA
Methylome-level assessment with broad genome coverage
Enhanced adapter ligation technology with NEXTFLEX® Ligation
Bead-based cleanup
Automation-friendly workflow is compatible with liquid handlers
Methylated barcoded adapters for multiplexing contain embedded index sequence
Functionally validated with Illumina® sequencing platforms
Kit Specs
| Cat # | Name | Quantity |
| NOVA-5119-01 | NEXTFLEX® Bisulfite Library Prep Kit for Illumina® Sequencing | 8 RXNS |
| NOVA-5119-02 | NEXTFLEX® Bisulfite Library Prep Kit for Illumina® Sequencing | 48 RXNS |
| NOVA-511911 | NEXTFLEX® BISULFITE-SEQ BARCODES - 6 | 48 RXNS |
| NOVA-511912 | NEXTFLEX® BISULFITE-SEQ BARCODES-12 | 96 RXNS |
| NOVA-511913 | NEXTFLEX® BISULFITE-SEQ BARCODES - 24 | 192 RXNS |
| NOVA-511921 | NEXTFLEX® Msp1 RESTRICTION ENZYME | 8 RXNS |
| NOVA-511922 | NEXTFLEX® Msp1 RESTRICTION ENZYME | 48 RXNS |
Protocol
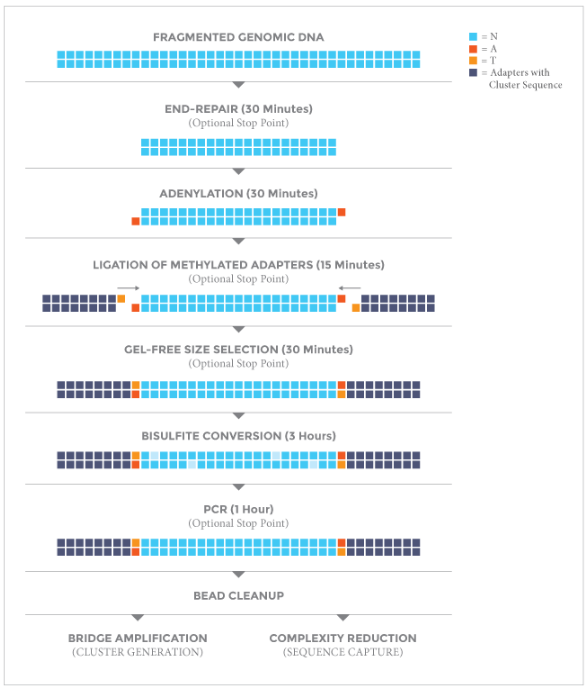
The NEXTFLEX® Bisulfite-Seq Library Prep Kit is designed for both reduced representation bisulfite sequencing (RRBS) and whole-genome bisulfite sequencing (WGBS). It generates single, paired-end and multiplexed DNA libraries for sequencing on Illumina® platforms. The NEXTFLEX® U+ Polymerase Mix included in the kit is able to read through uracil present in DNA templates and has been optimized to amplify bisulfite converted templates.
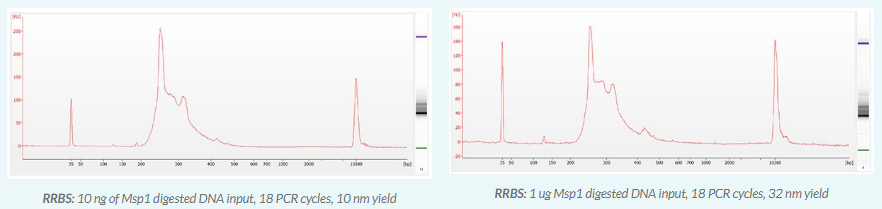
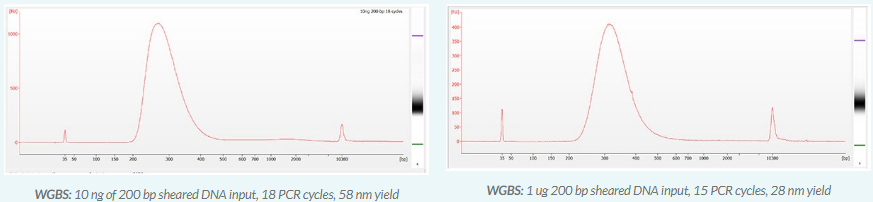
KIT CONTENTS
NEXTFLEX® Bisulfite-Seq End Repair Buffer Mix
NEXTFLEX® Bisulfite-Seq End Repair Enzyme Mix
NEXTFLEX® Bisulfite-Seq Adenylation Mix
NEXTFLEX® Bisulfite-Seq Ligation Mix
NEXTFLEX® Bisulfite-Seq DNA Adapter 24 (25 µM)
NEXTFLEX® Adapter Dilution Buffer
NEXTFLEX® Primer Mix (12.5 µM)
NEXTFLEX® Bisulfite-Seq U+ PCR Master Mix
Nuclease-free water
Resuspension Buffer
REQUIRED MATERIALS NOT PROVIDED
10 ng – 1 µg of fragmented /or/ Msp1 digested genomic DNA in up to 40 µL nuclease-free water.
NEXTFLEX® Bisulfite-Seq Barcodes – 6 / 12 / 24 (Cat # 511911, 511912, 511913 ) (Optional)
NEXTFLEX® Msp 1 – 48 reactions (Cat # 511922)
EZ DNA Methylation-Gold™ Kit (Zymo Research, Cat # D5005)
Ethanol 100% (room temperature)
Ethanol 80% (room temperature)
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) / or / similar
96 well Library Storage and Pooling Plate (Fisher Scientific, Cat # AB-0765) / or / similar
Adhesive PCR Plate Seal (BioRad, Cat # MSB1001)
Agencourt AMPure XP 5 mL (Beckman Coulter Genomics, Cat # A63880)
Magnetic Stand – 96 (Ambion, Cat # AM10027) / or / similar
Heat block
Thermocycler
2, 10, 20, 200 and 1000 µL pipettes / multichannel pipettes
Nuclease-free barrier pipette tips
Microcentrifuge
1.5 mL nuclease-free microcentrifuge tubes
Vortex
Papers that Cite the use of the NEXTFLEX® Bisulfite-Seq Kit:
Agarwal, P., et al. (2015) CGGBP1 mitigates cytosine methylation at repetitive DNA sequences. BMC Genomics, 16:390. doi:10.1186/s12864-015-1593-2.
Berg, A., et al. (2015) Obesity is Associated With DNA Methylation in Population-Based Adolescents. Circulation. Poster: 131:AP260, Session Title: Obesity.
Bewick, A., et. al. (2016) On the origin and evolutionary consequences of gene body DNA methylation. PNAS. 113: 9111 – 9116.
Ramesh, V., et. al. (2016) Loss of Uhrf1 in neural stem cells leads to activation of retroviral elements and delayed neurodegeneration. Genes & Development. 30: 2199 – 2212.
Satgé, C. et. al. (2016) Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nature Plants 2, 16166. doi:10.1038/nplants.2016.166.
Ye, R., et al. (2015) A Dicer-Independent Route for Biogenesis of siRNAs that Direct DNA Methylation in Arabidopsis. Molecular Cell. doi:10.1016/j.molcel.2015.11.015.








