
- 产品简介
- 工作流程
- 试剂组分
- 引用文献
NEXTflex™ 16S V1-V3 Amplicon-Seq Library Prep Kit 专为微生物16S核糖体RNA(rRNA)基因 V1-V3高变区文库制备设计的一款试剂盒,支持多重扩增子测序建库。兼容于Illumina Miseq测序平台双端测序,是监测微生物群落种群波动的理想实验方案。
低PCR偏差,高目标序列读取率实验流程
NEXTflex 16S V1-V3 Amplicon-Seq Kit实验流程中采用2步PCR,以增加样本特异性index,减少扩增子测序中的目标序列脱靶率。
产品特点:
快速-2小时文库制备
低样本起始量-只需1ng的基因组DNA
低PCR偏差,高目标序列读取率
测序引物都包含在此试剂盒中
多达384种接头用于多重测序
匹配PE的Sciclone NGS 和NGSx自动工作站
经Illumina MiSeq测序平台验证
产品列表:
| 货号 | 产品名称 | 规格 |
| NOVA-4202-02 | NEXTflex™ 16S V1-V3 Amplicon-Seq Kit (12 Barcodes) | 24 RXNS |
| NOVA-4202-03 | NEXTflex™ 16S V1-V3 Amplicon-Seq Kit (48 Barcodes) | 96 RXNS |
| NOVA-4202-04 | NEXTflex™ 16S V1-V3 Amplicon-Seq Kit (Barcodes 1-96) | 192 RXNS |
| NOVA-4202-05 | NEXTflex™ 16S V1-V3 Amplicon-Seq Kit (Barcodes 97-192) | 192 RXNS |
| NOVA-4202-06 | NEXTflex™ 16S V1-V3 Amplicon-Seq Kit (Barcodes 193-288) | 192 RXNS |
| NOVA-4202-07 | NEXTflex™ 16S V1-V3 Amplicon-Seq Kit (Barcodes 289-384) | 192 RXNS |
工作流程:
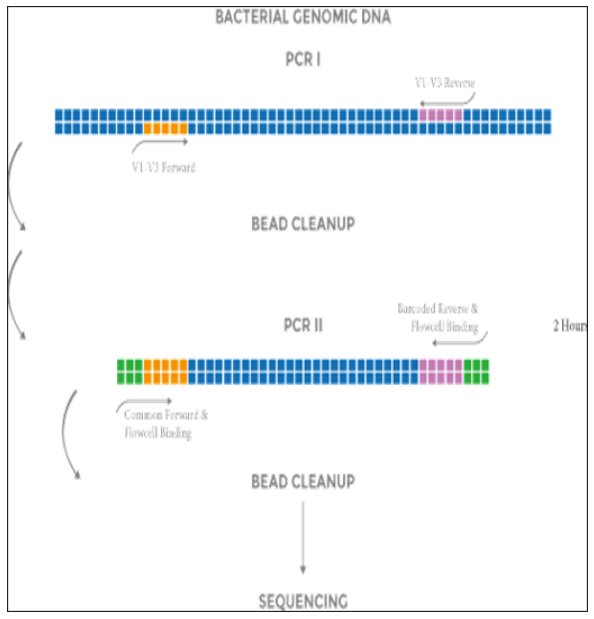
KIT CONTENTS
NEXTFLEX® PCR Master Mix
NEXTFLEX® 16S V1-V3 PCR I Primer Mix
NEXTFLEX® PCR II Barcoded Primer Mix
Resuspension Buffer
Nuclease-free Water
REQUIRED MATERIALS NOT PROVIDED
1 ng – 50 ng high-quality genomic DNA in up to 36 µL nuclease-free water for each library
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) or similar
Adhesive PCR Plate Seal (Bio-Rad®, Cat # MSB1001)
Agencourt® AMPure® XP 5 mL (Beckman Coulter® Genomics, Cat # A63880)
Magnetic Stand – 96 (Thermo Fisher® Scientific, Cat # AM10027) or similar
Thermocycler
2, 10, 20, 200 and 1000 µL pipettes / multichannel pipettes
Nuclease-free barrier pipette tips
Vortex
80% Ethanol, freshly prepared (room temperature)
NEXTflex 16S V1–V3 Amplicon-Seq Kit部分引用文献:
Cortez, V., Canal, E., Dupont-Turkowsky, J. C., Quevedo, T., Albujar, C., Chang, T., . . . Bausch, D. G. (2018). Identification of Leptospira and Bartonella among rodents collected across a habitat disturbance gradient along the Inter-Oceanic Highway in the southern Amazon Basin of Peru. Plos One, 13(10). doi:10.1371/journal.pone.0205068.
Franzen, J., Zirkel, A., Blake, J., Rath, B., Benes, V., Papantonis, A., & Wagner, W. (2016). Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells. Aging Cell, 16(1), 183-191. doi:10.1111/acel.12544.
Hakim, H., Dallas, R., Wolf, J., Tang, L., Schultz-Cherry, S., Darling, V., . . . Rosch, J. W. (2018). Gut Microbiome Composition Predicts Infection Risk During Chemotherapy in Children With Acute Lymphoblastic Leukemia. Clinical Infectious Diseases,67(4), 541-548. doi:10.1093/cid/ciy153.
Luna, R. A., Oezguen, N., Balderas, M., Venkatachalam, A., Runge, J. K., Versalovic, J., . . . Williams, K. C. (2017). Distinct Microbiome-Neuroimmune Signatures Correlate With Functional Abdominal Pain in Children With Autism Spectrum Disorder. Cellular and Molecular Gastroenterology and Hepatology, 3(2), 218-230. doi:10.1016/j.jcmgh.2016.11.008.
Maiuri, A. R., et al. (2017) Mismatch Repair Proteins Initiate Epigenetic Alterations during Inflammation-Driven Tumorigenesis. Cancer Research, 77(13), 3467-3478. doi:10.1158/0008-5472.can-17-0056.
Quereda, J. J., et al. (2016) Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. PNAS. doi:10.1073/pnas.1523899113.
Pahwa, R., Balderas, M., Jialal, I., Chen, X., Luna, R. A., & Devaraj, S. (2017). Gut Microbiome and Inflammation: A Study of Diabetic Inflammasome-Knockout Mice. Journal of Diabetes Research,2017, 1-5. doi:10.1155/2017/6519785.
Ranjan, R., Rani, A., Metwally, A., McGee, H. S. and Perkins D. L. (2015) Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophy Res Com. doi:10.1016/j.bbrc.2015.12.083.
Su, A., Yang, W., Zhao, L., Pei, F., Yuan, B., Zhong, L., . . . Hu, Q. (2018). Flammulina velutipes polysaccharides improve scopolamine-induced learning and memory impairment in mice by modulating gut microbiota composition. Food & Function, 9(3), 1424-1432. doi:10.1039/c7fo01991b.
Whon, T. W., Chung, W., Lim, M. Y., Song, E., Kim, P. S., Hyun, D., . . . Nam, Y. (2018). The effects of sequencing platforms on phylogenetic resolution in 16 S rRNA gene profiling of human feces. Scientific Data, 5, 180068. doi:10.1038/sdata.2018.68.
Yao, J. et al. (2016) A Pathogen-Selective Antibiotic Minimizes Disturbance to the Microbiome. Antimicrob. Agents Chemother. 00535-16. doi:10.1128/AAC.00535-16.








