
- 产品简介
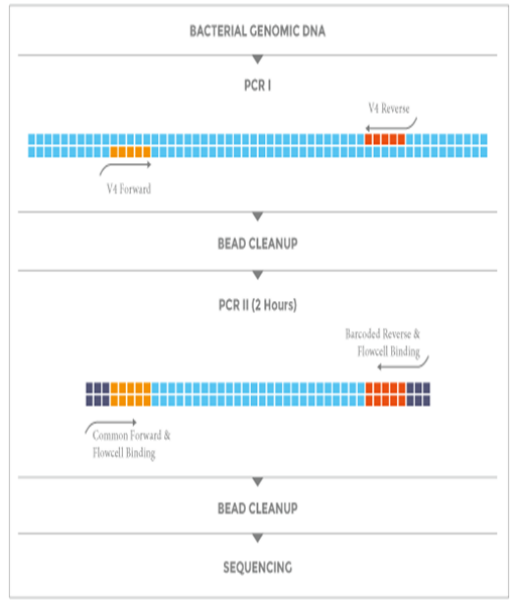
- 工作流程
- 试剂组分
- 引用文献
NEXTflex™ 16S V4 Amplicon-Seq Library Prep Kit 2.0 专为微生物16S核糖体RNA(rRNA)基因 V4高变区文库制备设计的一款试剂盒,支持多重扩增子测序建库。兼容于Illumina Miseq测序平台双端测序,是监测微生物群落种群波动的理想实验方案。

Figure 1. 使用NEXTflex™ 16S V4 Amplicon-Seq Library Prep Kit 2.0 建库,对16S V4 reads分析测量,样本为犬类粪便中提取的20ng DNA。
低PCR偏差,高目标序列读取率实验流程
NEXTflex 16S V4 Amplicon-Seq Kit 2.0 实验流程中采用2步PCR,以增加样本特异性index,减少扩增子测序中的目标序列脱靶率。
低样本起始量
NEXTflex 16S V4 Amplicon-Seq Library Prep Kit 2.0进行建库时所需样本量为 1 ng–50ng基因组DNA。
产品特点:
快速-2小时文库制备
简单-两步完成整个流程
低样本起始量-只需1ng的样品
低PCR偏差,高目标序列读取率
测序引物都包含在此试剂盒中
多达384种接头用于多重测序
匹配Sciclone NGS 和NGSx自动工作站
经Illumina测序平台验证
产品列表:
| 货号 | 产品名称 | 规格 |
| NOVA-4203-02 | NEXTflex™ 16S V4 Amplicon-Seq Kit 2.0 (12 Barcodes) | 24 RXNS |
| NOVA-4203-03 | NEXTflex™ 16S V4 Amplicon-Seq Kit 2.0 (48 Barcodes) | 96 RXNS |
| NOVA-4203-04 | NEXTflex™ 16S V4 Amplicon-Seq Kit 2.0(Barcodes 1-96) | 192 RXNS |
| NOVA-4203-05 | NEXTflex™ 16S V4 Amplicon-Seq Kit 2.0(Barcodes 97-192) | 192 RXNS |
| NOVA-4203-06 | NEXTflex™ 16S V4 Amplicon-Seq Kit 2.0(Barcodes 193-288) | 192 RXNS |
| NOVA-4203-07 | NEXTflex™ 16S V4 Amplicon-Seq Kit 2.0(Barcodes 289-384) | 192 RXNS |
工作流程:
KIT CONTENTS
NEXTFLEX® PCR Master Mix
NEXTFLEX® 16S V4 PCR I Primer Mix
NEXTFLEX® PCR II Barcoded Primer Mix
Resuspension Buffer
Nuclease-free Water
REQUIRED MATERIALS NOT PROVIDED
1 ng – 50 ng high-quality genomic DNA in up to 36 µL nuclease-free water for each library
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) or similar
Adhesive PCR Plate Seal (Bio-Rad®, Cat # MSB1001)
Agencourt® AMPure® XP 5 mL (Beckman Coulter® Genomics, Cat # A63880)
Magnetic Stand – 96 (Thermo Fisher® Scientific, Cat # AM10027) or similar
Thermocycler
2, 10, 20, 200 and 1000 µL pipettes / multichannel pipettes
Nuclease-free barrier pipette tips
Vortex
80% Ethanol, freshly prepared (room temperature)
NEXTflex 16S V4 Amplicon-Seq Kit 部分引用文献
Bekker, V., Zwittink, R. D., Knetsch, C. W., Sanders, I. M., Berghuis, D., Heidt, P. J., . . . Kuijper, E. J. (2019). Dynamics of the Gut Microbiota in Children Receiving Selective or Total Gut Decontamination Treatment During Hematopoietic Stem Cell Transplantation. Biology of Blood and Marrow Transplantation. doi:10.1016/j.bbmt.2019.01.037.
Bruce-Keller, A. J., Fernandez-Kim, S., Townsend, R. L., Kruger, C., Carmouche, R., Newman, S., . . . Berthoud, H. (2017). Maternal obese-type gut microbiota differentially impact cognition, anxiety and compulsive behavior in male and female offspring in mice. Plos One, 12(4). doi:10.1371/journal.pone.0175577.
Davey, M. P., Norman, L., Sterk, P., Huete-Ortega, M., Bunbury, F., Loh, B. K., . . . Smith, A. G. (2019). Snow algae communities in Antarctica – metabolic and taxonomic composition. New Phytologist. doi:10.1111/nph.15701.
Ford, S. L., Lohmann, P., Preidis, G. A., Gordon, P. S., Odonnell, A., Hagan, J., . . . Hair, A. B. (2019). Improved feeding tolerance and growth are linked to increased gut microbial community diversity in very-low-birth-weight infants fed mothers own milk compared with donor breast milk. The American Journal of Clinical Nutrition,109(4), 1088-1097. doi:10.1093/ajcn/nqz006.
Luk, B., Veeraragavan, S., Engevik, M., Balderas, M., Major, A., Runge, J., . . . Versalovic, J. (2018). Postnatal colonization with human “infant-type” Bifidobacterium species alters behavior of adult gnotobiotic mice. Plos One, 13(5). doi:10.1371/journal.pone.0196510.
Luna, R. A., Oezguen, N., Balderas, M., Venkatachalam, A., Runge, J. K., Versalovic, J., . . . Williams, K. C. (2017). Distinct Microbiome-Neuroimmune Signatures Correlate With Functional Abdominal Pain in Children With Autism Spectrum Disorder. Cellular and Molecular Gastroenterology and Hepatology, 3(2), 218-230. doi:10.1016/j.jcmgh.2016.11.008
Moon, C., Stupp, G. S., Su, A. I., & Wolan, D. W. (2017). Metaproteomics of colonic microbiota unveils discrete protein functions among colitic mice and control groups. doi:10.1101/219782.
Stalenhoef, J. E., et al. (2017) Fecal Microbiota Transfer for Multidrug-Resistant Gram-Negatives: A Clinical Success Combined With Microbiological Failure. Open Forum Infectious Diseases, 4(2). doi:10.1093/ofid/ofx047.
Svensson, K., Paruch, L., Gaby, J. C., & Linjordet, R. (2018). Feeding frequency influences process performance and microbial community composition in anaerobic digesters treating steam exploded food waste. Bioresource Technology, 269, 276-284. doi:10.1016/j.biortech.2018.08.096.
Sylvia, K. E., Deyoe, J. E., & Demas, G. E. (2018). Early-life sickness may predispose Siberian hamsters to behavioral changes following alterations of the gut microbiome in adulthood. Brain, Behavior, and Immunity, 73, 571-583. doi:10.1016/j.bbi.2018.07.001.
Whon, T. W., Chung, W., Lim, M. Y., Song, E., Kim, P. S., Hyun, D., . . . Nam, Y. (2018). The effects of sequencing platforms on phylogenetic resolution in 16 S rRNA gene profiling of human feces. Scientific Data, 5, 180068. doi:10.1038/sdata.2018.68.
Wotzkas, S. Y., Kreuzer, M., Maier, L., Zuend, M., Schlumberger, M., Nguyen, B., . . . Misselwitz, B. (2018). Microbiota stability in healthy individuals after single-dose lactulose challenge – a randomized controlled study. doi:10.1101/424531.
Zhao, R., Yang, W., Pei, F., Zhao, L., & Hu, Q. (2018). In vitro fermentation of six kinds of edible mushrooms and its effects on fecal microbiota composition. Lwt, 96, 627-635. doi:10.1016/j.lwt.2018.06.012.








