
NEXTFLEX® Rapid Directional RNA-Seq Kit 2.0
Reliable: High coverage uniformity with low duplication rate
Convenient: Optimized for use with 5 ng – 5 µg total RNA with reverse transcriptase and cleanup/size selection beads included
Efficient: Automated on the Sciclone® G3 NGSx and Zephyr® G3 NGS workstations
- Product description
- Protocol
- Data
- Kit Contents
New & Improved Library Preparation Kit for your RNA Sequencing Needs
The NEXTFLEX® rapid directional RNA-seq kit 2.0 produces libraries for Illumina® sequencing instruments with high coverage uniformity, low duplication rates, strand specificity and minimal rRNA contamination when used with the NEXTFLEX® Poly(A) Beads 2.0 (10 ng – 5 μg) or NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat) (5 ng – 1 μg). This kit includes reverse transcriptase, necessary library preparation reagents, and cleanup/size selection beads optimized to ensure reliable performance. The kit involves a simple library preparation protocol that has been validated with the updated NEXTFLEX® poly(A) beads 2.0 and NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat) to accommodate total RNA as input. The NEXTFLEX® rapid directional RNA-seq kit 2.0 is designed to be used with NEXTFLEX® RNA-Seq 2.0 Unique Dual Index Barcodes (6.25μM), which are color-balanced and have undergone proprietary purity QC metrics to generate reliable sequencing results for every sample.
Features
Reliable: High coverage uniformity with low duplication rate
Convenient: Optimized for use with 5 ng – 5 µg total RNA with reverse transcriptase and cleanup/size selection beads included
Streamlined: Simple protocol validated with NEXTFLEX® poly(A) beads 2.0 and NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat)
Flexible: Designed to work with NEXTFLEX® RNA-Seq 2.0 Unique Dual Index barcodes that allow a wide range of multiplexing (2 up to 1,526 samples in one run)
Efficient: Automated on the Sciclone® G3 NGSx and Zephyr® G3 NGS workstations
Kit Specs
| Cat # | Name | Quantity |
| NOVA-5198-01 | NEXTFLEX® Rapid Directional RNA-Seq Kit 2.0 | 8 RXNS |
| NOVA-5198-02 | NEXTFLEX® Rapid Directional RNA-Seq Kit 2.0 | 48 RXNS |
| NOVA-5198-03 | NEXTFLEX® Rapid Directional RNA-Seq Kit 2.0 | 96 RXNS |

PERFORMANCE OF POLY(A)-SELECTED LIBRARIES
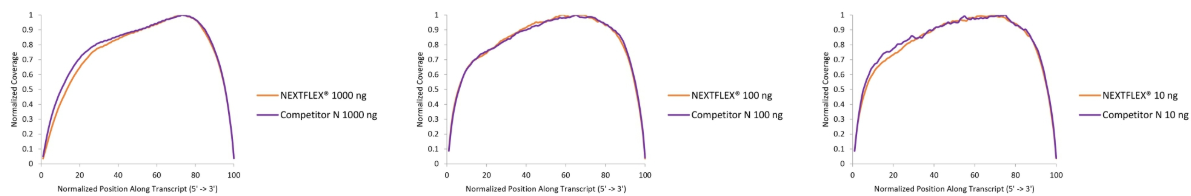
Figure 1. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrates even coverage along transcripts compared to the Competitor N kit. Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX® Poly(A) beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using paired-end mode (2×76 bp). Reads were trimmed using cutadapt and mapped to the Gencode v30 reference using bowtie2. The coverage along transcripts was calculated using the BBMap pileup tool.
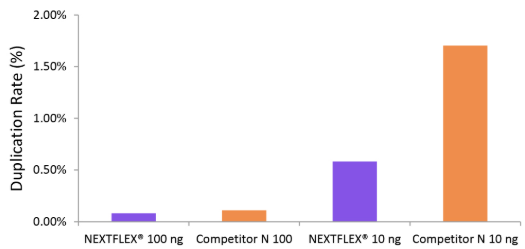
Figure 2. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrate low duplication rate compared to the Competitor N kit. Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX® Poly(A) beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using paired-end mode (2×76 bp). Reads were trimmed using cutadapt, mapped to the Gencode v30 reference using bowtie2, and randomly downsampled to 100k reads. Duplication rate was calculated using the fastp all-in-one FASTQ preprocessor.
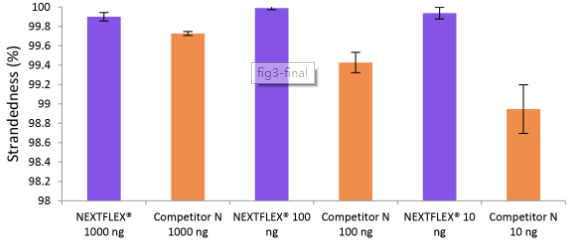
Figure 3. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrates comparable directionality to the Competitor N kit. Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) containing ERCC RNA Spike-In mix (Thermo Fisher Scientific #4456740) using the NEXTFLEX® Poly(A) Beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using paired-end mode (2×76 bp). Reads were trimmed using cutadapt and mapped to the ERCC92 reference using bowtie2. Strandedness was calculated using SAMtools.
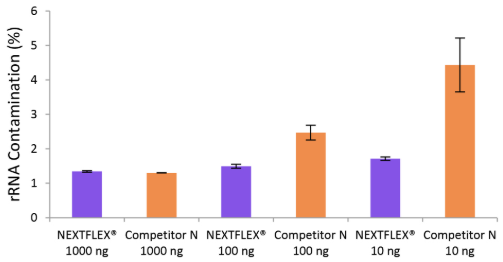
Figure 4. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 delivers libraries containing low levels of rRNA contamination than the Competitor N kit. Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX® Poly(A) Beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using paired-end mode (2×76 bp). The reads were trimmed using cutadapt and the percent of rRNA was determined by using bowtie2 to map reads to human rRNA. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrated superior removal of 5S, 5.8S, 12S, 16S, 18S, and 28S rRNA species compared to the Competitor N kit.
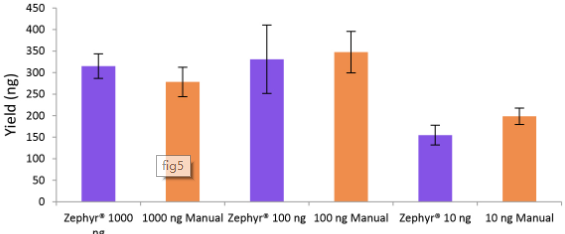
Figure 5. Libraries prepared using the Zephyr® G3 NGS workstation and manually deliver comparable yields using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0. Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent® #740000) using the NEXTFLEX® Poly(A) Beads 2.0. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0. Final library concentrations were quantified using the Qubit® 2.0 fluorometer (Thermo Fisher® Scientific #Q32866).
PERFORMANCE OF rRNA-DEPLETED LIBRARIES
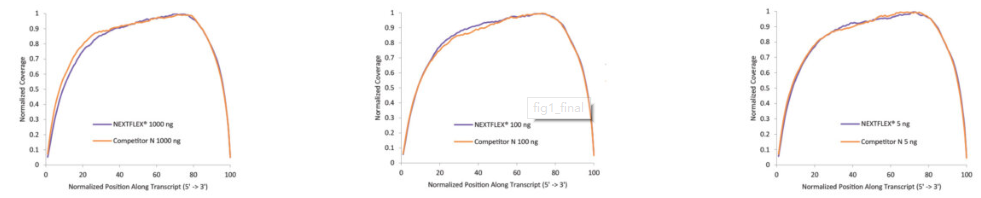
Figure 1. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrates even coverage along transcripts compared to the Competitor N kit. Poly(A) mRNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX® Poly(A) beads 2.0 and the Competitor N Poly(A) enrichment kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using paired-end mode (2×76 bp). Reads were trimmed using cutadapt and mapped to the Gencode v30 reference using bowtie2. The coverage along transcripts was calculated using the BBMap pileup tool.
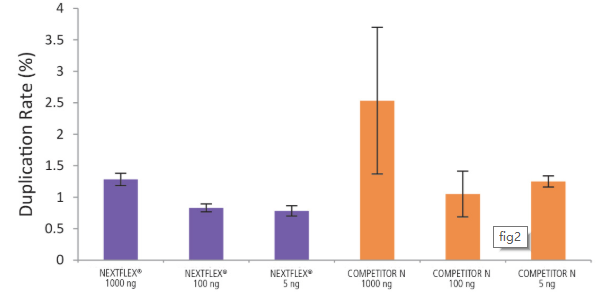
Figure 2. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrate low duplication rates compared to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent® #740000) using the NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using single-end mode (1×151 bp). Reads were trimmed using cutadapt, mapped to the Gencode v30 reference transcriptome using bowtie2, and randomly downsampled to 28k reads. Duplication rate was calculated using the fastp all-in-one FASTQ preprocessor.
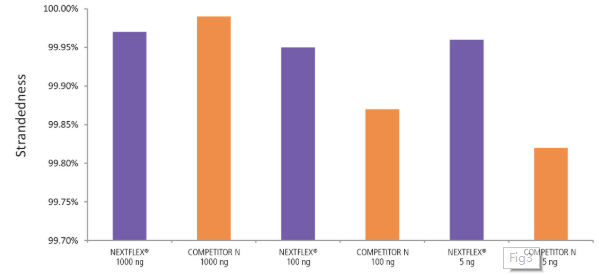
Figure 3. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrates comparable directionality relative to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent® #740000) using the NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using single-end mode (1×151 bp). Reads were trimmed using cutadapt and mapped to the Gencode v30 reference transcriptome using bowtie2. Reads from respective samples were combined and downsampled for a total of 800k reads each. Strandedness was calculated using the fastp all-in-one FASTQ preprocessor.
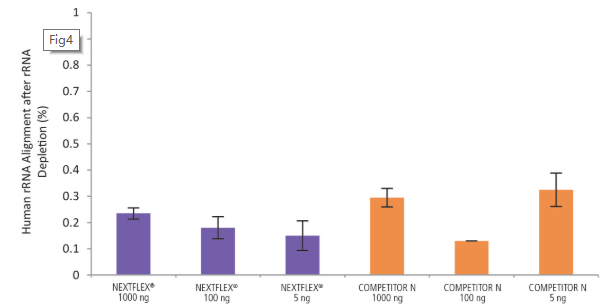
Figure 4. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 delivers libraries containing low levels of rRNA contamination compared to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using single-end mode (1×151 bp). The reads were trimmed using cutadapt and the percent of rRNA was determined by using bowtie2 to map reads to human rRNA. The NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 demonstrated superior removal of 5S, 5.8S, 12S, 16S, 18S, and 28S rRNA species compared to the Competitor N kit.
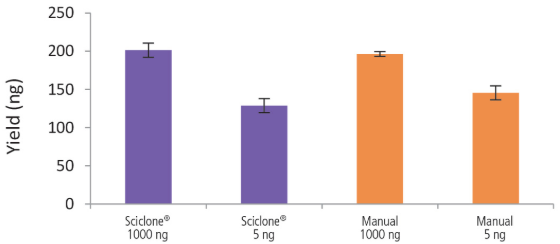
Figure 5. Libraries prepared using the Sciclone® G3 NGSx workstation and manually deliver comparable yields using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX® RiboNaut™ rRNA depletion kit. Libraries were generated using the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0. Final library concentrations were quantified using the Qubit® 2.0 fluorometer (Thermo Fisher Scientific #Q32866).
KIT CONTENTS
NEXTFLEX® RNA-Seq Fragmentation Buffer 2.0
NEXTFLEX® Directional First Strand Synthesis Buffer Mix 2.0
NEXTFLEX® Rapid Reverse Transcriptase 2.0
NEXTFLEX® Directional Second Strand Synthesis Mix 2.0
NEXTFLEX® RNA-Seq Adenylation Buffer Mix 2.0
NEXTFLEX® RNA-Seq Adenylation Enzyme 2.0
NEXTFLEX® RNA-Seq Ligase Buffer Mix 2.0
NEXTFLEX® RNA-Seq Ligase Enzyme 2.0
NEXTFLEX® RNA-Seq PCR Master Mix 2.0
NEXTFLEX® RNA-Seq Primer Mix 2.0 (50 µM)
Nuclease-free Water
Resuspension Buffer
NEXTFLEX® Cleanup Beads XP
REQUIRED MATERIALS NOT PROVIDED
10 ng – 1 µg total RNA for enrichment by NEXTFLEX® Poly(A) Beads or ~ 1 ng – 100 ng isolated mRNA or 5 ng – 1 µg rRNA-depleted total RNA using NEXTFLEX® RiboNaut™ rRNA depletion kit (human, mouse, rat) or 1 ng – 100 ng of isolated mRNA or rRNA-depleted RNA using alternative methods
DynaMag™-2 Magnet (Thermo Fisher Scientific, Cat # 123-21D)
100% Ethanol (stored at room temperature)
2, 10, 20, 200 and 1000 µL pipettes
RNase-free pipette tips
Nuclease-free 1.5 mL microcentrifuge tubes
Thin wall nuclease-free 0.5 mL microcentrifuge tubes
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) / or / similar
Adhesive PCR Plate Seal (Bio-Rad, Cat # MSB1001)
Magnetic Stand -96 (Thermo Fisher Scientific, Cat # AM10027) / or / similar for post PCR cleanup
Microcentrifuge
Thermal cycler
Heat block
Vortex








