
NEXTFLEX® Rapid Directional RNA-Seq Library Prep Kit
Robust, reliable performance
Up to 96 barcodes available for multiplexing
Minimal hands-on time required
- Product description
- Data
- Kit Contents
- Citations
NEXTFLEX® Rapid Directional RNA-Seq Library Prep Kit
The NEXTFLEX® Rapid Directional RNA-Seq Kit provides an easy and flexible solution for generating single read, paired-end and multiplexed stranded libraries from mRNA or rRNA depleted total RNA. This kit can be used to obtain “stranded” information which identifies the specific DNA strands a given RNA transcript was derived from with >99% strand specificity. This stranded information improves transcript annotation and improves read alignment, reducing per sample sequencing costs. dUTP-based strand orientation also enables the detection of antisense expression regulatory relationships and allows for the discovery of novel ncRNA.
Cost Effective, Stranded RNA Library Prep Kit Bundles
To simplify your mRNA sequencing, PerkinElmer now offers affordable bundles containing the NEXTFLEX® Rapid Directional RNA-Seq library prep reagents, NEXTFLEX® RNA-Seq Barcodes and NEXTFLEX® Poly(A) Beads. There are two different versions of this kit. They each contain the 48 rxn NEXTFLEX® Rapid Directional RNA-Seq Kit, NEXTFLEX® Poly(A) Beads (48 rxns) and 24 unique NEXTFLEX® RNA-Seq Barcodes (in aliquots of 2 reactions each).
Flexible Multiplexing Options for Illumina® RNA Sequencing
The NEXTFLEX® Rapid Directional RNA-Seq Library Prep Kit was designed to be used in conjunction with the NEXTFLEX® RNA-Seq Barcodes or NEXTFLEX-96™ RNA-Seq Barcodes for multiplexing. These single index barcodes are available in sets of 6, 12, 24, 48 and 96 barcodes.
Poly(A) Beads for mRNA Purification
The NEXTFLEX® Poly(A) Beads 2.0 offer a convenient method, validated with the NEXTFLEX® Rapid Directional RNA-Seq Kit, for purification of pure, intact mRNA.
Features
Explore our NEW! NEXTFLEX® Rapid Directional RNA-Seq Kit 2.0 for an updated chemistry that delivers high performance.
2 hours faster than traditional Illumina® stranded RNA-Seq library prep protocols
Provides precise measurement of strand orientation (>99%)
>10 ng – 1 µg total RNA for enrichment by NEXTFLEX® Poly(A) Beads 2.0 or ~ 1 ng – 100 ng isolated mRNA or rRNA-depleted RNA
Minimal hands-on time required
Streamlined protocol reduces sample loss
Utilizes dUTP-based methodology
Robust, reliable performance
Up to 96 barcodes available for multiplexing
Automation protocols are available for the Sciclone® NGS and NGSx workstations
Functionally validated with Illumina® sequencing platforms
Allows for single read or paired end transcriptome sequencing
Kit Specs
| Cat # | Name | Quantity |
| NOVA-5138-07 | NEXTflex™ Rapid Directional RNA-Seq Kit | 8 RXNS |
| NOVA-5138-08 | NEXTflex™ Rapid Directional RNA-Seq Kit | 48 RXNS |
| NOVA-5138-10 | NEXTflex™ W/ RNA-SEQ BARCODES 1 - 24 & POLY(A) BEADS | 48 RXNS |
| NOVA-5138-11 | NEXTflex™ W/ RNA-SEQ BARCODES 25 - 48 & POLY(A) BEADS | 48 RXNS |
Streamlined Library Construction for Quantitative, Directional, and Standard RNA-Seq
RNA Sequencing (RNA-Seq) is a valuable tool for a broad range of clinical, environmental, and basic research. Producing high quality RNA-Seq libraries can be challenging for several reasons, including isolation of pure RNA, efficiently converting RNA to cDNA, and loss of material incurred during the series of enzymatic steps and cleanups required for library construction. Here we introduce PerkinElmer’s family of NEXTFLEX® Rapid RNA-Seq kits for Illumina® sequencing, all of which include the thermostable NEXTFLEX® Rapid Reverse Transcriptase enzyme. The NEXTFLEX® Rapid RNA-Seq Kits provide affordable and unique technology, some of the shortest RNA-Seq library preparation times of any kits on the market, include all enzymes required for library preparation, and produce the highest quality RNA-Seq library.
Introduction
The NEXTFLEX® Rapid RNA-Seq Kits allow the end user to complete library construction in less than 1 day. In contrast, the previous generation of NEXTFLEX® RNA-Seq Kits, as well as the current Illumina® TruSeq® RNA-Seq Kits, require over 7 hours for completion (Table 1). By combining second strand synthesis and end repair into the same reaction, the need for a separate end repair step and cleanup is eliminated. This improvement is time efficient and reduces loss of material during cleanup.
Table 1. Comparison of RNA-Seq library preparation kit protocols and time to completion. Note that times include bead cleanups and account for time required from RNA fragmentation to the final bead cleanup after PCR.
| Kit | Time to Completion | Steps Required | Bead Cleanups Required | Reverse Transcriptase Included? |
| NEXTFLEX® Rapid RNA-Seq Kit | 5 hrs 50 mins | 6 | 3 | Yes |
| Illumina® TruSeq® RNA V2 | 7 hrs 10 mins | 7 | 4 | No |
Thermostable NEXTFLEX® Rapid Reverse Transcriptase for Improved First Strand Synthesis and Library Yield
A critical step in RNA-Seq library construction is the conversion of RNA to cDNA. All NEXTFLEX® Rapid RNA-Seq Kits include NEXTFLEX® Rapid Reverse Transcriptase (RT) enzyme, in contrast to our previous RNA-Seq kits as well as to the Illumina® TruSeq® kit (Table 1). The NEXTFLEX® Rapid RT is a thermostable, RNaseH minus enzyme that functions optimally at 50˚ C, a higher temperature than standard Moloney-Murine Leukemia Virus (M-MLV) reverse transcriptases, which function at 42˚ C. This elevated temperature allows for reduced secondary structure in RNA templates and therefore increased efficiency of first strand synthesis.
To examine the efficacy of NEXTFLEX® Rapid RT in library construction, we compared library yields using cDNA produced by different enzymes. The same pool of Poly (A)+ selected mRNA isolated from murine Ag8 cells was used in first strand synthesis with either NEXTFLEX® Rapid RT or SuperScript® III (Figure 1). Higher library yields were obtained using the NEXTFLEX® Rapid RT coupled with the NEXTFLEX® Rapid RNA-Seq Kit. These results demonstrate improved library yields as a result of optimized first strand synthesis using NEXTFLEX® Rapid RT.
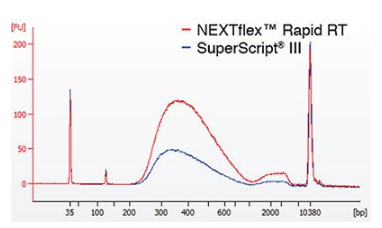
Figure 1. Improved library yield using NEXTFLEX® Rapid RT. High Sensitivity DNA Bioanalyzer traces of RNA-Seq libraries constructed with NEXTFLEX® Rapid RNA-Seq Kits. Libraries were constructed using 10 ng of fragmented, Poly (A)+ mRNA converted to cDNA using either NEXTFLEX® Rapid RT (blue) or SuperScript® III (red).
High-quality RNA-Seq Data
Further analysis of NEXTFLEX® Rapid RNA-Seq library quality was carried out using Illumina® sequencing, as library yield is only one metric of library quality. Libraries were prepared with the NEXTFLEX® Rapid RNA-Seq Kit using 10 ng aliquots of a single murine Ag8 cell Poly (A)+ RNA sample, so as to disentangle biological variation from technical variation. Libraries were sequenced on an Illumina® HiSeq® 2500 instrument using a 67 bp single end RAPID run. Resulting reads were trimmed based on a quality score moving window using sickle and mapped to the UCSC mm10 assembly using TopHat 2.0.10. We obtained a total of 31,949,336 reads for Rapid RT libraries and 31,720,922 for Superscript III libraries (Table 2). For transcript representation purposes, only reads mapping to exons, specifically all annotated 5’ UTRs, coding sequences, and 3’ UTRs, were further considered. The number of total reads as well as unique reads mapping to exons were similar between the two enzymes; however, a greater number of transcripts, 12,418 vs. 12,129, were represented in the NEXTflex Rapid RT libraries vs. SuperScript III libraries, respectively.
Table 2. Read counts in NEXTFLEX® Rapid RNA-Seq libraries and unique reads after mapping to exons.
| Rt Enzyme | NEXTFLEX® Rapid RT | Superscript® III RT |
| Total Reads | 31,949,336 | 31,720,922 |
| Reads After Quality Trimming | 31,891,608 | 31,663,364 |
| Unique Reads Mapping to Exons | 17,411,368 | 17,487,086 |
| Unique Reads in Exons (%) | 54.60 | 55.23 |
| Number Transcripts Represented | 12,418 | 12,129 |
A further analysis of mapped reads was performed. Read quality before and after mapping was visualized using FastQC and GC content as a function of reads examined (Figure 2). Read quality is excellent in the total library and is slightly higher in the remaining set of trimmed, mapped reads, which is expected given a quality-aware trimming step was carried out. Furthermore, GC content is very similar to the theoretical distribution, indicating accuracy in transcript representation. Visual inspection of several genes demonstrates read coverage across all exons (Figure 3A and B). Furthermore, a metagene analysis of read coverage across all genes divided into 100 bin segments, demonstrates even read coverage across the 5’-, gene body, and 3’-ends of transcripts (Figure 3C).
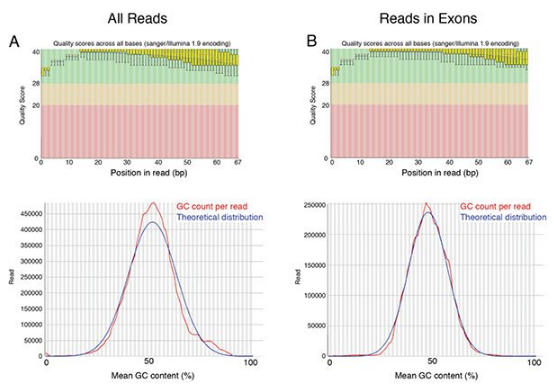
Figure 2. Metrics of NEXTFLEX® Rapid RNA-Seq data determined by FastQC. Quality score plots for (top) and mean GC content (bottom) for (A) all reads or (B) reads mapping to exons. Plots shown correspond to NEXTFLEX® Rapid RNA-Seq libraries constructed using NEXTFLEX® Rapid RT.
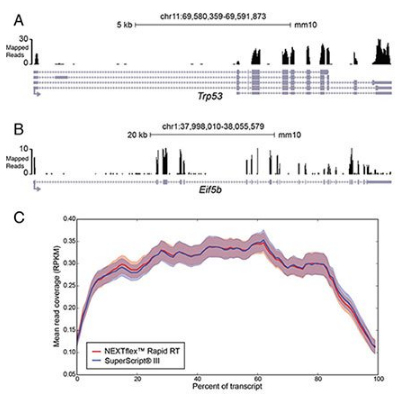
Figure 3. Read coverage across gene bodies. Mapped reads at the (A) Trp53 and (B) Eif5b loci scaled to read density as indicated. (C) Metagene plot of read density across all annotated loci. All gene bodies and mapped read densities are scaled to 100 bin segments; mean read density is shown in reads per kilobase per million mapped reads (RPKM; solid line) +/- standard error across replicates (faded bands). Shown is read signal corresponding to libraries made with either NEXTFLEX® Rapid RT (red) or SuperScript III (blue).
Finally, we examined library consistency across protocols. We compared RNA-Seq read counts in exons from libraries constructed using the NEXTFLEX® Rapid RNA-Seq Kit to longer traditional protocols. Indeed, libraries constructed using the two methods were highly similar (r = 0.9976; Figure 4). These data demonstrate that the NEXTFLEX® Rapid RNA-Seq protocol provides significantly faster library construction with enhanced reverse transcriptase performance.
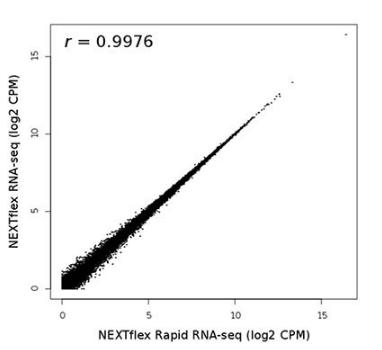
Figure 4. Correlation between Rapid and Standard RNA-Seq data. Pearson’s correlation between the average log2 of normalized counts per million (CPM) of all genes across all replicates of Rapid RNA-Seq vs. Standard RNA-Seq, r = 0.9976.
Conclusions
The present study demonstrates the utility of the NEXTFLEX® Rapid RNA-Seq Kits as a method to improve speed of library preparation without any compromise in library quality. All NEXTFLEX® Rapid RNA-Seq Kits include NEXTFLEX® Rapid RT, a high performance thermostable enzyme, thus providing all components required for library construction within a single kit. NEXTFLEX® Rapid RNA-Seq Kits produced high-quality sequencing data with improved transcript representation. In addition, the increased library yield using NEXTFLEX® Rapid RT is an important improvement for users with low-input amounts. For researchers wishing to multiplex libraries, PerkinElmer continues to offer our full range of up to 96 NEXTFLEX® RNA-Seq adapter barcodes. The NEXTFLEX® Rapid RNA-Seq and Rapid Directional RNA-Seq kits provide a streamlined workflow for users to create high-quality RNA-Seq libraries with less hands-on time.
5’ to 3’ Sequence Coverage
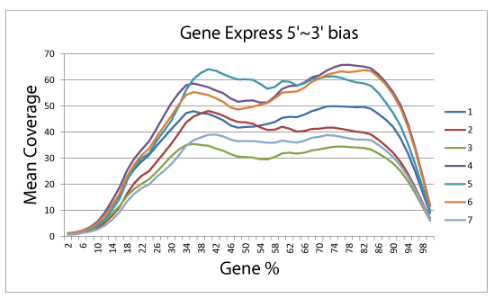
Normalized coverage by position. For each library, the average coverage is shown at each relative position along the transcripts’ length.
KIT CONTENTS
NEXTFLEX® RNA Fragmentation Buffer
NEXTFLEX® First Strand Synthesis Primer
NEXTFLEX® Directional First Strand Synthesis Buffer Mix
NEXTFLEX® Rapid Reverse Transcriptase
NEXTFLEX® Directional Second Strand Synthesis Mix
NEXTFLEX® Adenylation Mix
NEXTFLEX® Ligation Mix
NEXTFLEX® RNA-Seq Barcode Adapter 1 (0.6 µM)
NEXTFLEX® Uracil DNA Glycosylase
NEXTFLEX® Primer Mix (12.5 µM)
NEXTFLEX® PCR Master Mix
Nuclease-free Water
Resuspension Buffer
REQUIRED MATERIALS NOT PROVIDED
10 ng – 1 µg total RNA for enrichment by NEXTFLEX® Poly(A) Beads 2.0 or ~ 1 ng – 100 ng isolated mRNA or rRNA-depleted RNA
DynaMag™-2 Magnet (Life Technologies Cat # 123-21D)
Commercial kits for rRNA depletion (optional, if not using poly(A) enrichment)
100% Ethanol (stored at room temperature)
80% Ethanol ( freshly prepared )
2, 10, 20, 200 and 1000 µL pipettes
RNase-free pipette tips
Nuclease-free 1.5 mL microcentrifuge tubes
Thin wall nuclease-free 0.5 mL microcentrifuge tubes
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) / or / similar
Adhesive PCR Plate Seal (BioRad, Cat # MSB1001)
Agencourt AMPure XP 60 mL (Beckman Coulter Genomics, Cat # A63881)
Magnetic Stand -96 (Ambion, Cat # AM10027) / or / similar for post PCR cleanup
Microcentrifuge
Thermocycler
Heat block
Vortex
Selected Publications that Reference Using the NEXTFLEX® Rapid Directional RNA Library Prep Kit:
Beyaz, S. et al. (2016) High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 531, 53–58. doi:10.1038/nature17173.
Blance, S., et al. (2016) Stem cell function and stress response are controlled by protein synthesis. Stem cell function and stress response are controlled by protein synthesis. Nature, 534, 335–340. doi:10.1038/nature18282.
Bonizzoni, M., et al. (2013) Probing functional polymorphisms in the dengue vector, Aedes aegypti. BMC Genomics. 14:739 http://www.biomedcentral.com/1471-2164/14/739.
Carnes, M. U., et al. (2015) The Genomic Basis of Postponed Senescence in Drosophila melanogaster. PLoS ONE. doi: 10.1371/journal.pone.0138569.
Denzler, R., Agarwal V., Stefano J., Bartel DP. and Stoffel, M. (2014) Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 54:4 pp 766-76.
Dharshinia, S., et al. (2016) De novo sequencing and transcriptome analysis of a low temperature tolerant Saccharum spontaneum clone IND 00-1037. J of Biotechnology. doi: 10.1016/j.jbiotec.2016.05.036.
Dobáková, E., Flegontov, P., Skalický, T. and Lukeš, J. (2015) Unexpectedly Streamlined Mitochondrial Genome of the Euglenozoan Euglena gracilis. Genome Biol Evol. 7. 3358-3367. doi:10.1093/gbe/evv229.
Fang, W. and Bartel, D. P. (2015) The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Molecular Cell. 60:1. p131–145. doi: 10.1016/j.molcel.2015.08.015.
Guérin, F., Isnard, C., Cattoir, V. and Giard, J. C. (2015) Complex Regulation Pathways of AmpC-Mediated β-Lactam Resistance in Enterobacter cloacae Complex. Antimicrob. Agents Chemother., 59: 7753 – 7761. doi: 10.1128/AAC.01729-15.
Jones, B. M., Wcislo, W. T. and Robinson, G. E. (2015) Developmental Transcriptome for a Facultatively Eusocial Bee, Megalopta genalis. g3, Oct 2015; 5: 2127 – 2135. doi: g3.115.021261v1.
Lamanna, F., et al. (2015) Cross-tissue and cross-species analysis of gene expression in skeletal muscle and electric organ of African weakly-electric fish (Teleostei; Mormyridae). BMC Genomics. 16:668 . doi:10.1186/s12864-015-1858-9.
Lin, M.-H., Jones, D. F. and Fleming, R. (2015) Transcriptomic analysis of degraded forensic body fluids, Forensic Science International: Genetics, Volume 17. 35-42. dio: 10.1016/j.fsigen.2015.03.005.
McNeill, M. S., Kapheim, K. M., Brockmann, A., McGill, T. A. W., Robinson, G. E. (2015) Brain regions and molecular pathways responding to food reward type and value in honey bees. Genes, Brain and Behavior. doi: 10.1111/gbb.12275.
Mullenders, J., et al. (2015) Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 2015; 212:1833-1850. doi: 10.1084/jem.20151323.
Nam, J., Rissland, O.S., Koppstein, D. et al. (2014) Global Analyses of the Effect of Different Cellular Contexts on MicroRNA Targeting. Molecular Cell. http://dx.doi.org/10.1016/j.molcel.2014.02.013.
Pham, K. T. M., et al. (2015) MoSET1 (Histone H3K4 Methyltransferase in Magnaporthe oryzae) Regulates Global Gene Expression during Infection-Related Morphogenesis. PLOS Genetics. doi: 10.1371/journal.pgen.1005385.
Rittschofa, C. C., et al. (2014) Neuromolecular responses to social challenge: Common mechanisms across mouse, stickleback fish, and honey bee. PNAS. doi: 10.1073/pnas.1420369111.
Rossetto CC, Tarrant-Elorza M, Pari GS (2013) Cis and Trans Acting Factors Involved in Human Cytomegalovirus Experimental and Natural Latent Infection of CD14 (+) Monocytes and CD34 (+) Cells. PLoS Pathog 9(5): e1003366. doi:10.1371/journal.ppat.1003366.
Rube, H. T., et al. (2016) Sequence features accurately predict genome-wide MeCP2 binding in vivo. Nature Communications. 7:11025. doi:10.1038/ncomms11025.
Solovchenko, A., et al. (2016) Nitrogen availability modulates CO2 tolerance in a symbiotic chlorophyte, Algal Research, 16, 177-188. doi: 10.1016/j.algal.2016.03.002.
Tarvin, R. D., Santos, J. C., O’Connell, L. A., Zakon, H. H. and Cannatella, D. C. (2016) Convergent Substitutions in a Sodium Channel Suggest Multiple Origins of Toxin Resistance in Poison Frogs. Mol Biol Evol. 33:4. 1068-1081. doi: 10.1093/molbev/msv350.
Van Laar, T, A., Chen, T, You, T., and Leung, K. P. (2015) Sublethal Concentrations of Carbapenems Alter Cell Morphology and Genomic Expression of Klebsiella pneumoniae Biofilms. Antimicrobial Agents and Chemotherapy. doi: 10.1128/AAC.04581-14.
Wheeler, M. M. and Robinson G. E. (2014) Diet-dependent gene expression in honey bees: honey vs. sucrose or high fructose corn syrup. Scientific Reports. 4: 5726. doi:10.1038/srep05726.
Zhang, Y. et al. (2015) Genome-, Transcriptome- and Proteome-Wide Analyses of the Gliadin Gene Families in Triticum urartu. PLOS One. doi: 10.1371/journal.pone.0131559.








