
NEXTFLEX® Rapid DNA-Seq Kit 2.0
Complete library prep solution, including size selection beads
Bundle kits available with library prep reagents and barcodes
Up to 384 barcodes available upon request
- Product description
- Protocol
- Data
- Kit Contents
- Citations
NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms
The NEXTFLEX® rapid DNA-seq 2.0 kit is designed to prepare multiplexed libraries for single or paired-end sequencing using Illumina® platforms. The enhanced NEXTFLEX® kit simplifies workflow by using master mixed reagents and magnetic bead based cleanup, reducing pipetting and eliminating time-consuming steps in library preparation. The NEXTFLEX® rapid DNA-seq kit 2.0 produces library preps with a larger number of unique sequencing reads, allowing for robust genome coverage in challenging GC-rich sequences that can introduce bias.
The NEXTFLEX® rapid DNA-seq 2.0 kit bundle offers all the advantages and reagents of the NEXTFLEX Rapid DNA-seq kit 2.0 and includes NEXTFLEX-HT™ barcodes (8, 48, or 96 reaction size kits) supplied in a single-use format. This allows the lab to tie all the necessary components of the library prep workflow to a single lot for ease of purchasing and tracking of consumables.
Features
Complete library prep solution, including size selection beads
Bundle kits available with library prep reagents and barcodes
Up to 384 barcodes available upon request
Convenient and cost-effective package in a 8, 48, or 96 reaction format
Simplified workflow
Automation compatible with liquid handlers, including the PerkinElmer Sciclone® G3 NGS/NGSx workstations and Zephyr® G3 NGS workstation
Robust genome coverage
Reliable performance with GC-bias and sequencing bias mitigation
Low input requirement for genomic DNA down to 1 ng
For FFPE samples, use the NEXTFLEX® Rapid XP DNA-Seq Kit
Functionally validated with Illumina® sequencing platforms
Kit Specs
| Cat # | Name | Quantity |
| NOVA-5188-01 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | (NO BARCODES) 8 RXNS |
| NOVA-5188-02 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | (NO BARCODES) 48 RXNS |
| NOVA-5188-03 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | (NO BARCODES)96 RXNS |
| NOVA-5188-11 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | BARCODES 1-8 8 RXNS |
| NOVA-5188-12 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | BARCODES 1-48 48 RXNS |
| NOVA-5188-13 | NEXTFLEX® Rapid DNA-Seq Kit 2.0 for Illumina® Platforms | BARCODES 1-96 96 RXNS |
Protocol
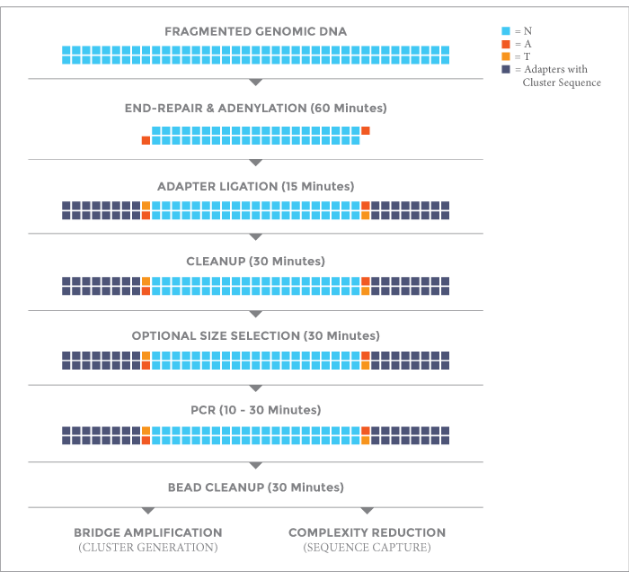
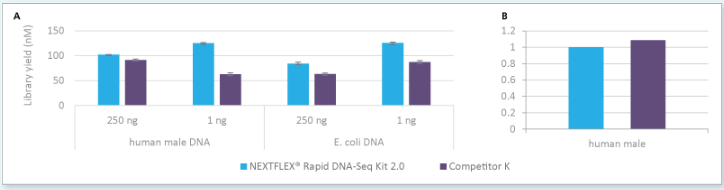
Figure 1: Libraries prepared with NEXTFLEX® rapid DNA-Seq kit 2.0 show higher yield than libraries prepared with Competitor K’s kit using the same number of PCR cycles from a 250 ng and 1 ng input (A) and robust conversion rate (B).
Additional Data between NEXTFLEX® Rapid DNA-Seq 2.0 and Competitor K’s Kit Libraries
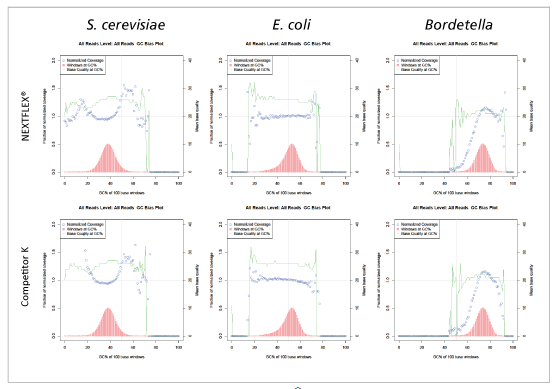
Figure 2: Libraries prepared with NEXTFLEX® Rapid DNA-seq 2.0 shows less GC bias than libraries prepared with Competitor K’s kit.
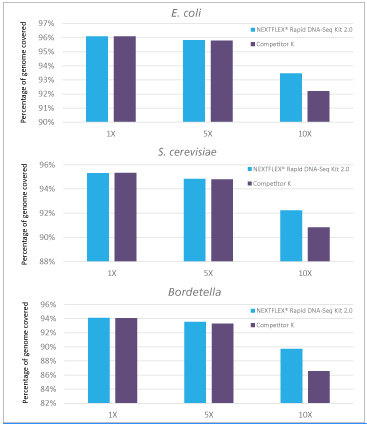
Figure 3: Libraries prepared with NEXTFLEX® Rapid DNA-seq 2.0 shows higher genome coverage than libraries prepared with Competitor K’s kit.
KIT CONTENTS
NEXTFLEX® End Repair & Adenylation Buffer Mix 2.0
NEXTFLEX® End Repair & Adenylation Enzyme Mix 2.0
NEXTFLEX® Ligase Buffer Mix 2.0
NEXTFLEX® Ligase Enzyme 2.0
NEXTFLEX® PCR Master Mix 2.0
NEXTFLEX® Primer Mix 2.0
NEXTFLEX® Cleanup Beads 2.0
Nuclease-free Water
Resuspension Buffer
REQUIRED MATERIALS NOT PROVIDED
1 ng – 1 µg of fragmented genomic DNA in up to 32 µL nuclease-free water.
NEXTflex® barcodes if purchasing a non-bundled kit (includes NEXTflex-HT™ barcodes)
Ethanol 100% (room temperature)
Ethanol 80% (room temperature)
Covaris System (S2, E210) or other method for DNA fragmentation
96 well PCR Plate Non-skirted (Phenix Research, Cat # MPS-499) or similar
96 well Library Storage and Pooling Plate (Thermo Fisher Scientific, Cat # AB-0765) or similar
Adhesive PCR Plate Seal (Bio-Rad, Cat # MSB1001)
Magnetic Stand -96 (Thermo Fisher Scientific, Cat # AM10027) or similar
Heat block
Thermocycler
2, 10, 20, 200 and 1000 µL pipettes / multichannel pipettes
Nuclease-free barrier pipette tips
Microcentrifuge
1.5 mL nuclease-free microcentrifuge tubes
Vortex
Shain, A. H., et al. (2015) The Genetic Evolution of Melanoma from Precursor Lesions. The New England Journal of Medicine. doi: 10.1056/NEJMoa1502583.
Campbella, M. A., et al. (2015) Genome expansion via lineage splitting and genome reduction in the cicada endosymbiont Hodgkinia. PNAS. doi: 10.1073/pnas.1421386112.
Gal, C. et. al. (2015) The impact of the HIRA histone chaperone upon global nucleosome architecture. Cell Cycle. 14:1, 123-134. doi:10.4161/15384101.2014.967123.
Yang, Y. A., et al. (2016) FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Research. doi: 10.1093/nar/gkw498.
Palomo, A., et al. (2016) Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. The ISME Journal. doi:10.1038/ismej.2016.63.
Brodie, J., et al. (2016) Characterising the microbiome of Corallina officinalis, a dominant calcified intertidal red alga. FEMS Microbiology Ecology.doi:10.1093/femsec/fiw110.
Givnish, T. J., et al. (2016) Phylogenomics and historical biogeography of the monocot order Liliales: out of Australia and through Antarctica. Cladistics. doi: 10.1111/cla.12153.
Harrisson, K., et al. (2016) Pleistocene divergence across a mountain range and the influence of selection on mitogenome evolution in threatened Australian freshwater cod species. Heredity. doi:10.1038/hdy.2016.8.








