
LabChip® GX Touch™ Nucleic Acid Analyzer
Quantitate DNA and RNA sample integrity hours faster than agarose gels
Data visualization options (electropherogram, virtual gel, or tabular report)
NGS library prep analysis and QC
- Product description
- Data
- LabChip Assays
- Citations
DNA and RNA quantitation and sizing can be done in seconds using automated capillary electrophoresis separation. The LabChip® GX Touch™ nucleic acid analyzer’s microfluidic technology generates reproducible, high-resolution data and is optimal for:
Fast, high-throughput QC analysis of NGS libraries for detection of mutations associated with SARS-CoV-2
NGS library preparation (smear and fragment analysis) and quality control
RNA and DNA fragment analysis (including cell-free DNA, DNA isolated from FFPE samples, and PCR-free libraries)
Quantitation and qualification for CRISPR fragment analysis
Screening nucleic acid source material for SARS-CoV-2 vaccine development
Characterization and QC of small RNA molecules and CRISPR/Cas9 gRNA
Multiplex Detection of SARS-CoV-2 Multiplex Detection of SARS-CoV-2 Variants of Concern using ARMS-PCR
The LabChip GX Touch nucleic acid analyzer also supports a variety of DNA and RNA analysis, which provides unparallel genomic data quality with minimal input requirements. When the concentrations are low, and samples are precious, The LabChip® DNA High Sens and RNA Pico assays are ideal choice for reliable measurement and sample preservation. With complete analysis of genomic material in about 30 seconds, the LabChip GX Touch nucleic acid analyzer eliminates the nucleic acid quantitation workflow bottleneck. High-resolution analysis provides exact sizing and quantitation of DNA fragments and smears down to 5 bp from samples as small as 2 pg/µL.
The system offers a broad variety of DNA analysis options, including the NGS 3K assay, which provides unparalleled genomic data quality with minimal input requirements. This high degree of sensitivity also makes it the ideal instrument for gDNA, cfDNA, and PCR-free DNA quantitation, where concentrations are low, samples are precious and their preservation is essential.
Purity and Integrity Analysis of mRNA-based Vaccines
By using LabChip® RNA Assay, researchers at Merck developed an assay to characterize purity and integrity of mRNA vaccines. This fast, easy and efficient workflow helped speed up vaccine development and meet increased manufacturing high volume demand.
Features
The Comprehensive Solution for Genomic Sizing & Quantitation
Quantitate DNA and RNA sample integrity hours faster than agarose gels
Maximize rare or low-concentration samples like cfDNA, PCR-free libraries, and ChIP-seq libraries
Accurate quantitation down to 25 pg/µL for DNA smears and 0.5 pg/µL for fragments
NGS library prep analysis and QC
Flexible throughput options make it cost-efficient – 96-well or 384-well (HT) format
Data visualization options (electropherogram, virtual gel, or tabular report)
Applications
Complete NGS workflows
CRISPR Fragment Analysis
Vaccine Development
Specifications
| Dimensions | |
| Height | 69 cm | 27 in |
| Width | 51 cm | 20 in |
| Depth | 49 cm | 19.5 in |
Windows 10-compatible software
Increasing Speed and Throughput of SARS-CoV-2 Variant Detection Workflows
The capacity and speed of analysis make the LabChip® NGS 3K assay run on the LabChip® GX Touch™ nucleic acid analyzer an ideal component of high-throughput NGS-based variant detection workflows. Figure 1 illustrates examples of fully constructed library of CoV+ sample with pooled amplicon input at 200 ng (blue line) and 1200 ng (red line). It only takes a minute to examine NEXTFLEX® Variant Seq™ SARS-CoV-2 libraries, to ensure proper library sizing quality and exclusion of contaminating fragments.
Minimal sample consumption to save constructed library for next sequencing steps
Automated process for high measurement reproducibility and operation reliability
Multiple digitized data formats for convenient data analysis, visualization, and sharing
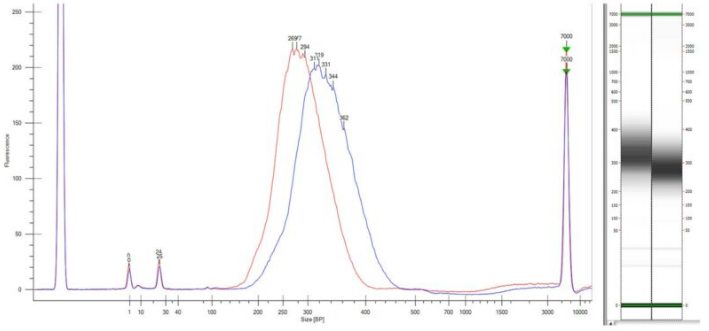
Figure 1: Validation of NEXTFLEX® Variant Seq™ SARS-CoV-2 Libraries.
Libraries of CoV+ samples were analyzed using the LabChip® NGS 3K assay on the LabChip® GX Touch™ nucleic acid analyzer: 200 ng pooled amplicon input (blue) and 1200 ng pooled amplicon input (red).
| Assay | Reagent Kit | 24 Chip | HT Chip |
| DNA 1K | CLS760673 | CLS138948 | 760517 |
| DNA 5K | CLS760675 | CLS138949 | 760435 |
| DNA 12K | 760569 | CLS138948 | 760517 |
| DNA High Sens | CLS760672 | CLS138948 | 760517 |
| Genomic DNA | CLS760685 | CLS138948 | 760517 |
| DNA NGS 3K | CLS960013 | CLS145331 | CLS144006 |
| RNA | CLS960010 | CLS138949 | 760435 |
| RNA Pico | CLS960012 | CLS138949 | 760435 |
| Small RNA | CLS153530 | CLS138949 | 760435 |
The system offers a broad variety of DNA analysis options, including the NGS 3K assay, which provides unparalleled genomic data quality with minimal input requirements. This high degree of sensitivity also makes it the ideal instrument for gDNA, cfDNA, and PCR-free DNA quantitation, where concentrations are low, samples are precious, and their preservation is essential.
Bagheri, H., Friedman, H., Shao, H., Chong, Y., Lo, C., Emran, F., . . . Peterson, A. (2018). TIE: A Method to Electroporate Long DNA Templates into Preimplantation Embryos for CRISPR-Cas9 Gene Editing. The CRISPR Journal,1(3), 223-229. doi:10.1089/crispr.2017.0020.
Chesnais, V., Ott, A., Chaplais, E., Gabillard, S., Pallares, D., Vauloup-Fellous, C., . . . Ginoux, E. (2018). Using massively parallel shotgun sequencing of maternal plasmatic cell-free DNA for cytomegalovirus DNA detection during pregnancy: A proof of concept study. Scientific Reports, 8(1). doi:10.1038/s41598-018-22414-6.
Costa, J-M., et al. (2018). Cell-free fetal DNA versus maternal serum screening for trisomy 21 in pregnant women with and without assisted reproduction technology: a prospective interventional study. Genetics in Medicine. doi:10.1038/gim.2018.4.
Faiz, A., Heijink, I. H., Vermeulen, C. J., Guryev, V., Berge, M. V., Nawijn, M. C., & Pouwels, S. D. (2018). Cigarette smoke exposure decreases CFLAR expression in the bronchial epithelium, augmenting susceptibility for lung epithelial cell death and DAMP release. Scientific Reports,8(1). doi:10.1038/s41598-018-30602-7.
Ford, L., Carter, G. P., Wang, Q., Seemann, T., Sintchenko, V., Glass, K., . . . Kirk, M. D. (2018). Incorporating Whole-Genome Sequencing into Public Health Surveillance: Lessons from Prospective Sequencing of Salmonella Typhimurium in Australia. Foodborne Pathogens and Disease, 15(3), 161-167. doi:10.1089/fpd.2017.2352.
Gergen, J., Coulon, F., Creneguy, A., Elain-Duret, N., Gutierrez, A., Pinkenburg, O., . . . Haspot, F. (2018). Multiplex CRISPR/Cas9 system impairs HCMV replication by excising an essential viral gene. Plos One, 13(2). doi:10.1371/journal.pone.0192602.
Giand et al. (2021) Early transmission of SARS-CoV-2 in South Africa: An epidemiological and phylogenetic report. International Journal of Infectious Diseases, 103, 234-241, doi.org/10.1016/j.ijid.2020.11.128.
Kim, K.W., et al. (2021) Respiratory viral co-infections among SARS-CoV-2 cases confirmed by virome capture sequencing. Sci Rep 11, 3934 doi.org/10.1038/s41598-021-83642-x.
Mogilevsky, M., Shimshon, O., Kumar, S., Mogilevsky, A., Keshet, E., Yavin, E., Heyd, F., and Karni, R. (2018) Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. doi: 10.1093/nar/gky921.
Ong, J., Woldhuis, R. R., Boudewijn, I. M., Berg, A. V., Kluiver, J., Kok, K., . . . Brandsma, C. A. (2019). Age-related gene and miRNA expression changes in airways of healthy individuals. Scientific Reports,9(1). doi:10.1038/s41598-019-39873-0.
Raffaele, J., Loughney, J. W., & Rustandi, R. R. (2021). Development of a microchip capillary electrophoresis method for determination of the purity and integrity of mrna in lipid nanoparticle vaccines. ELECTROPHORESIS. https://doi.org/10.1002/elps.202100272.
Trolle, C., et al. (2016) Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Scientific Reports (6). doi:10.1038/srep34220.








