
NEXTFLEX® Rapid XP DNA-Seq Kit
Reliable: Consistent yields and high-quality sequencing metrics
Streamlined: Single step for fragmentation, end-repair, and adenylation with minimal hands-on time
Flexible: Up to 1,536 color-balanced NEXTFLEX® barcodes, allowing a wide range of multiplexing, are available separately
- Product description
- Protocol
- Data
- Kit Contents
The NEXTFLEX® Rapid XP DNA-Seq kit combines enzymatic fragmentation with end-repair and A-tailing in one reaction to create a highly efficient first step in library generation, which is followed by ligation and PCR (optional for PCR-free workflows). This effective one-tube workflow produces DNA-seq libraries with consistent library size, high yield, low GC-bias, and high coverage.
Simplify Library Construction with Pre-Plated NEXTFLEX® Rapid XP DNA-seq Kits
Pre-plated NEXTFLEX® library prep kits eliminate the need for plate set-up, offering preprogrammed library construction on the Sciclone® G3 NGS/NGSx, Sciclone® G3 NGSx iQ™ and Zephyr® G3 NGS workstations. Pre-plated kits allow labs to:
Save time spent on manual set-up and reduce overall library prep time
Reduce the chance for human error
Remove inefficiencies due to dead volume
Flexible Multiplexing Options with 1,536 Barcodes Available for High-Throughput Multiplexing
We offer a broad range of color-balanced NEXTFLEX® adapters that improve multiplexing capabilities for both low-level and high-level multiplexing needs. A set of 1,536 NEXTFLEX® UDI adapters which are compatible with the NEXTFLEX® Rapid XP DNA-seq kit are now available for ultra high-throughput applications.
The NEXTFLEX® Rapid XP DNA-seq kit is also compatible with NEXTFLEX® DNA Barcodes, NEXTFLEX-96™ DNA Barcodes, NEXTFLEX® ChIP-Seq Barcodes, and NEXTFLEX-HT™ barcodes.
Barcodes are not supplied with the NEXTFLEX® Rapid XP DNA-seq kit and need to be purchased separately.
Features
Reliable: Consistent yields and high-quality sequencing metrics
Convenient: Kit includes fragmentation enzyme, cleanup/size selection beads, and conditioning solution to improve the fragmentation efficiency for DNA stored in buffers containing EDTA
Streamlined: Single step for fragmentation, end-repair, and adenylation with minimal hands-on time
Flexible: Up to 1,536 color-balanced NEXTFLEX® barcodes, allowing a wide range of multiplexing, are available separately
Efficient: Pre-plated version available for easier automation on the Sciclone® G3 NGS/NGSx and the Sciclone® G3 NGSx iQ™ workstations
Kit Specs
| Cat # | Name | Quantity |
| NOVA-5149-01 | NEXTFLEX® Rapid XP DNA-Seq Kit for Illumina® Platforms | 8 RXNS |
| NOVA-5149-02 | NEXTFLEX® Rapid XP DNA-Seq Kit for Illumina® Platforms | 48 RXNS |
| NOVA-5149-03 | NEXTFLEX® Rapid XP DNA-Seq Kit for Illumina® Platforms | 96 RXNS |
| NOVA-5149-103 | NEXTFLEX® Rapid XP DNA-Seq Kit for Illumina® Platforms(PRE-PLATED) | 96 RXNS |
Alternative FFPE Input Protocol
The NEXTFLEX® rapid XP DNA-seq kit protocol has been performance-validated using quality gDNA. The kit can also accommodate FFPE samples using an alternative protocol. Note that the user should verify quality of FFPE material prior to use with the kit. In order to prepare a library from FFPE samples, the following basic FFPE protocol can be followed.
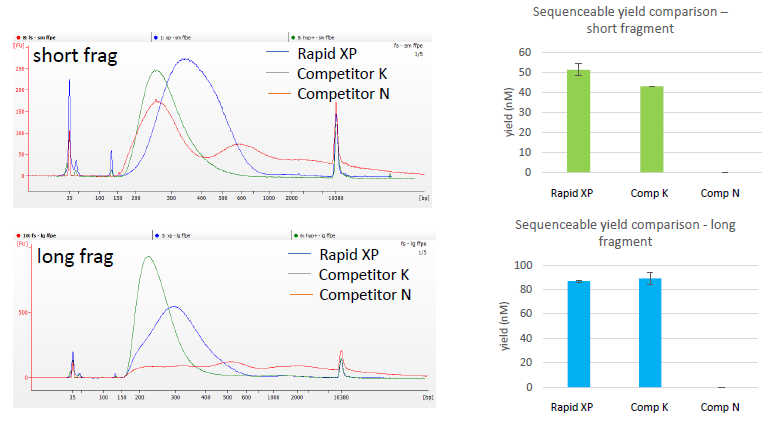
Figure 1: Library trace for 50 ng FFPE sample input into the NEXTFLEX® rapid XP DNA-seq kit using 24 minutes fragmentation time. TOP – short fragment FFPE, BOTTOM – long fragment FFPE sample.
Alternative Low Input Protocol
The NEXTFLEX® rapid XP DNA-seq kit protocol has been performance-validated using quality gDNA down to 1 ng. The kit can also accommodate low input sample down to 100 pg using an alternative protocol. Note that the user should verify quality of starting material prior to use with the kit. In order to prepare a library from low input samples, the following basic low input protocol can be followed.
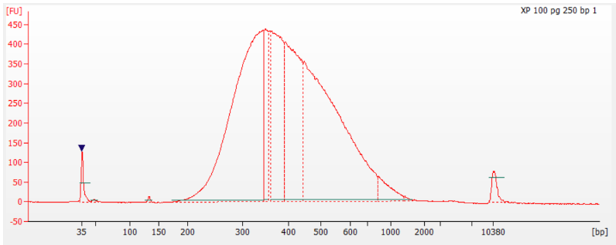
Figure 2: Library trace for low-input 100 pg of gDNA into the NEXTFLEX® rapid XP DNA-seq kit using 50 minutes fragmentation time.
Streamlined Enzymatic Fragmentation and Library Prep Workflow for Illumina® Sequencing
The NEXTFLEX® Rapid XP DNA-Seq kit combines enzymatic fragmentation with end-repair and A-tailing in one reaction to create a highly efficient first step in library generation, which is followed by ligation and PCR (optional for PCR-free workflows). This effective one-tube workflow produces DNA-seq libraries with consistent library size, high yield, low GC-bias, and high coverage.
The kit includes all the required reagents for fragmentation, library prep, and magnetic bead-based cleanup, and it is compatible with your NEXTFLEX® barcodes of choice. This DNA-seq library prep kit is highly flexible in terms of sample requirements, accommodating a range gDNA input amounts from 1 ng to 1 µg to quickly and efficiently generate high-quality libraries for Illumina® sequencing.
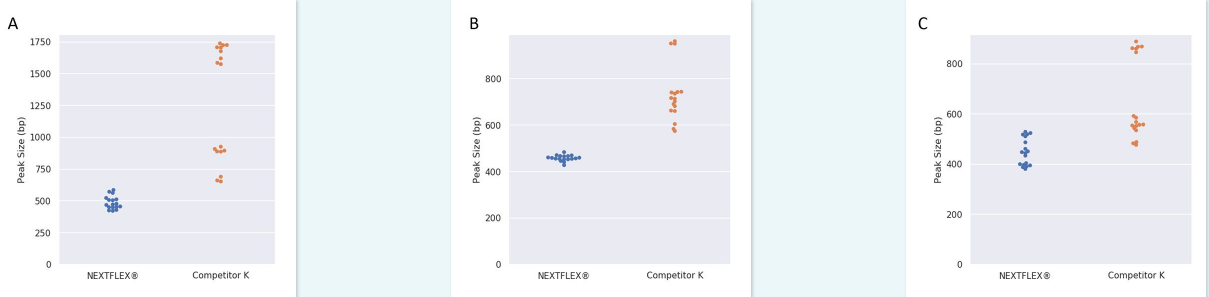
Figure 1: Reliable performance of NEXTFLEX® Rapid XP DNA-seq fragmentation enzyme. DNA is fragmented more evenly and into smaller sizes using the NEXTFLEX® Rapid XP DNA-seq kit than Competitor K’s kit. Each dot indicates each library peak size. (A) Libraries were prepared from 1 µg of DNA with 7 minutes of fragmentation with 2 PCR cycles: NEXTFLEX® Rapid XP DNA-seq kit (484 ± 52 bp, 11 %, n=17), Competitor K’s kit (1266 ± 451 bp, 36 %, n=17). (B) Libraries were prepared from 100 ng of DNA with 8 minutes of fragmentation with 5 PCR cycles: NEXTFLEX® Rapid XP DNA-seq kit (457 ± 12 bp, 3 %, n=18), Competitor K’s kit (730 ± 119 bp, 16 %, n=17). (C) Libraries were prepared from 1 ng of DNA with 15 minutes of fragmentation with 12 PCR cycles: NEXTFLEX® Rapid XP DNA-seq kit (448 ± 54 bp, 12 %, n=17), Competitor K’s kit (628 ± 160 bp, 25 %, n=17). All different input libraries were generated from 6 different commercially available DNA samples. (mean ± standard deviation, coefficient of variation)
Conditioning Solution included with the kit for DNA samples stored in EDTA
EDTA reduces the efficiency of enzymatic fragmentation. Since DNA is frequently eluted and stored in TE (10 mM Tris, 1 mM EDTA) or other buffers containing EDTA, the NEXTFLEX® Rapid XP DNA-Seq kit now includes a Conditioning Solution to improve the fragmentation efficiency of these DNA samples without disrupting the workflow.
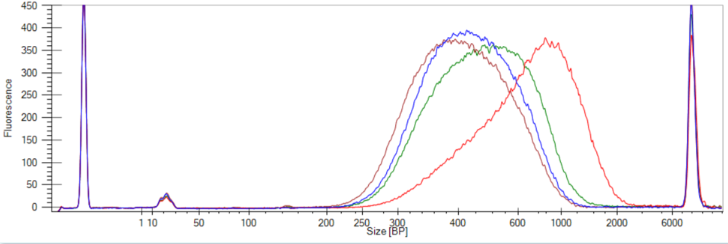
Figure 2: Libraries prepared from 100 ng of E. coli genomic DNA using the NEXTFLEX® Rapid XP DNA-seq kit and fragmented for 25 minutes with the NEXTFLEX® fragmentation enzyme. No EDTA/no Conditioning Solution (Blue), 1 mM EDTA/no Conditioning Solution (Red), No EDTA/1 μL Conditioning Solution (Brown), and 1 mM EDTA/1 μL Conditioning Solution (Green).
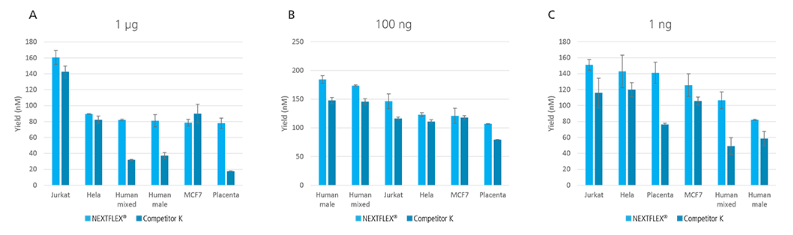
Figure 3: High yield obtained for libraries prepared using the NEXTFLEX® Rapid XP DNA-seq kit. Libraries were prepared using both NEXTFLEX® Rapid XP DNA-seq kit and Competitor K’s kit. (A) Libraries were prepared from 1 µg of DNA with 7 minutes of fragmentation with 2 PCR cycles. (B) Libraries were prepared from 100 ng of DNA with 8 minutes of fragmentation with 5 PCR cycles. (C) Libraries were prepared from 1 ng of DNA with 15 minutes of fragmentation with 12 PCR cycles.
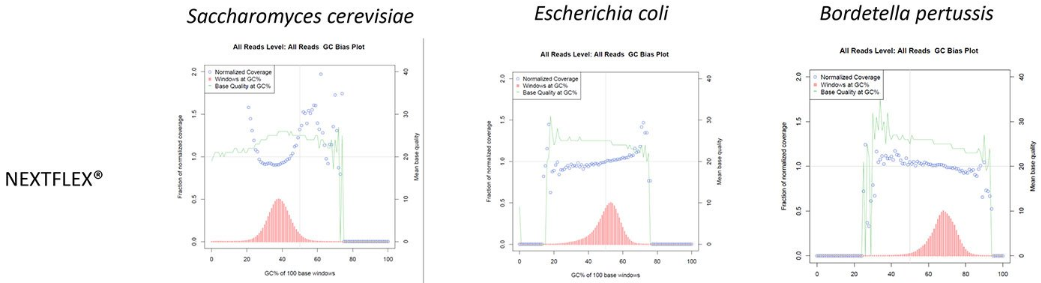

Figure 4. Libraries prepared with the NEXTFLEX® Rapid XP DNA-seq kit show less or comparable GC-bias than libraries prepared with Competitor K’s kit. Picard GC bias analysis, which show read coverage over the indicated genome with respect to GC content, is shown as a fraction of normalized coverage (blue) calculated in 100 bp windows (red) and plotted against the left y-axis. Mean base quality at each window (green) is calculated and plotted against the right y-axis.
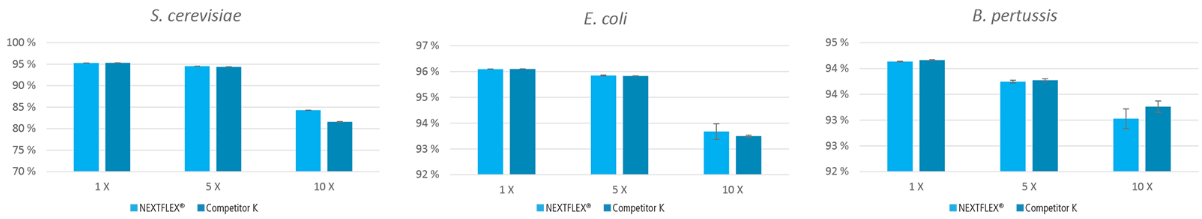
Figure 5. Libraries prepared with the NEXTFLEX® Rapid XP DNA-seq kit show higher or comparable genome coverage compared to libraries prepared using Competitor K’s kit. The percentage of genome bases covered at a minimum depth of 1X, 5X, and 10X of S. cerevisiae, E. coli, and B. pertussis.
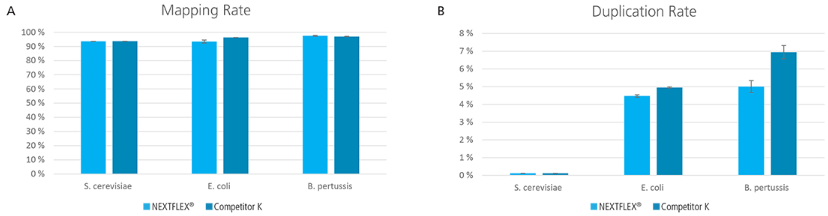
Figure 6. Libraries prepared with the NEXTFLEX® Rapid XP DNA-seq kit resulted in better or comparable data with respect to mapping rate and duplication rate, compared to libraries prepared using Competitor K’s kit. (A) Mapping rates represented as the percentage of reads which aligned to the indicated genome. (B) Duplication rates represented as the percentage of reads predicted to be PCR duplicates by Picard MarkDuplicates.
KIT CONTENTS
NEXTFLEX® Fragmentation Buffer
NEXTFLEX® Fragmentation Enzyme Mix
NEXTFLEX® Ligase Buffer Mix XP
NEXTFLEX® Ligase Enzyme XP
NEXTFLEX® PCR Master Mix XP
NEXTFLEX® Primer Mix XP
NEXTFLEX® Cleanup Beads XP
Conditioning Solution
Nuclease-free Water
Resuspension Buffer
REQUIRED MATERIALS NOT PROVIDED
1 ng – 1 µg of DNA in up to 34 µL nuclease-free water.
NEXTFLEX® barcodes
Ethanol 80% (room temperature)
96 well PCR Plate Non-skirted (Phenix® Research, Cat # MPS-499) or similar
96 well Library Storage and Pooling Plate (Thermo Fisher® Scientific, Cat # AB-0765) or similar
Adhesive PCR Plate Seal (Bio-Rad®, Cat # MSB1001)
Magnetic Stand -96 (Thermo Fisher® Scientific, Cat # AM10027) or similar
Thermocycler
2, 10, 20, 200 and 1000 µL pipettes / multichannel pipettes
Nuclease-free barrier pipette tips
Vortex








