
myBaits Expert –Plant–Angiosperms353
Predesigned Panel – Designed by expert plant geneticists
Proven results – Demonstrated utility across broad taxonomic range
Freely accessible pipelines – Simple, user-friendly data analysis
- Product description
- Data
- Publications
This new panel enriches 100's of single-copy genes orthologous across all angiosperms (flowering plants).
Combining the flexible hybridization power of in-solution target capture with an expertly selected set of orthologous locus sequences, this new probe set has been demonstrated to enrich hundreds of putatively single-copy protein-coding genes across a broad range of angiosperms (flowering plants). Probes were designed from 353 loci, each with 5-15 representative sequences from across all angiosperms which were selected using a novel “k-medoids clustering approach” to maximize taxonomic breadth of the design (Johnson, Pokorny, Dodsworth et al 2018, Systematic Biology).
These probes are broadly applicable for phylogenetic research across all flowering plants. For example, see the latest release of the Kew Tree of Life Explorer where users can access DNA sequence data and explore evolutionary relationships between thousands of samples covering over half of all angiosperm genera and nearly all families, generated using the Angiosperms353 probes. The kit is available from Daicel Arbor Biosciences as an in-stock catalog kit available for immediate shipment at a low per-reaction cost. As with all myBaits kits, the Angiosperms353 panel is provided as a complete solution target capture kit, including buffers, blockers, and baits, along with an easy-to-use protocol.
Predesigned Panel – Designed by expert plant geneticists
Proven results – Demonstrated utility across broad taxonomic range
Freely accessible pipelines – Simple, user-friendly data analysis
Cost-effective – Use one bait set across multiple distantly-related taxa
Kit Specs
| Ordering Information | Size | Cat# |
| myBaits Expert Angiosperms 353 v1 | 8 Reactions | 308108.v5 |
| 48 Reactions | 308148.v5 | |
| 96 Reactions | 308196.v5 |
Enrich Hundreds of Angiosperm Exons and their Flanking Regions
For hundreds of years, scientists have worked to unravel the complex evolutionary origins of the “angiosperms” group of land plants. This enormous, diverse group includes many economically important and ecologically interesting species, and the exact genetic relationships between these groups are still unresolved.
As the power and efficiency of genomic sequencing has increased in recent years, researchers have continued to innovate methods to tackle these major questions. One such method is the Angiosperms353 panel, which uses the flexible, versatile power of NGS hybridization capture to enrich for hundreds of conserved exons believed to be present across the angiosperms group. This universal tool to study exons and their flanking intronic regions allows phylogenomics and population genetics researchers to efficiently focus their sequencing efforts on informative regions that are directly comparable across datasets, for maximum research utility.
A Universal Phylogenomics Solution for Flowering Plants
The myBaits Expert Angiosperms353 kit has been demonstrated to work across a very wide taxonomic and sample range, with excellent exonic and intronic recovery that varies depending on the species and the quality of DNA input. Some example data are shown below, but for the latest and most comprehensive results, check out the most recent Publications that have been published utilizing this kit for an even greater variety of species and sample types.
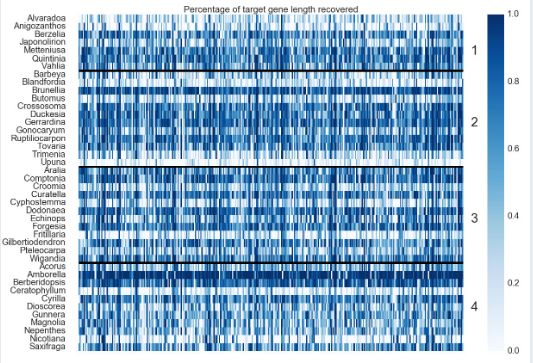
Fig 1. “Heatmap of Gene Recovery Efficiency. Each row is one sample, and each column is one gene. Colors indicate the percentage of the target length (calculated by the mean length of all k-medoid transcripts for each gene) recovered.” (pg 19, Johnson, Pokorny, Dodsworth et al 2019, Syst Bio). Image is Figure 3 from Johnson, Pokorny, Dodsworth et al 2019, Syst Biol.
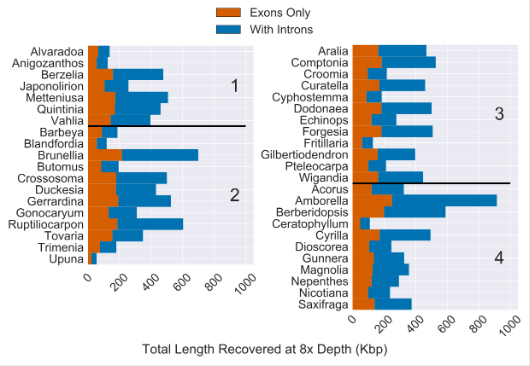
Fig 2. “Total Length of Sequence Recovery for Both Coding and Non-coding Regions Across 353 Loci for 42 Angiosperm Species. Reads were mapped back to either coding sequence (yellow) or coding sequence plus flanking non-coding (i.e. intron) sequence (purple)… The total length of coding sequence targeted was 260,802 bp. The median recovery of coding sequence was 137,046 bp and the median amount of non-coding sequence recovered was 216,816 bp (with at least 8x depth of coverage).” (pg 21, Johnson, Pokorny, Dodsworth et al 2019, Syst Bio). Image is modified version of Figure 4 from Johnson, Pokorny, Dodsworth et al 2019, Syst Bio.
Featured Publications
The publications listed below highlight some uses of the myBaits® Expert Angiosperms353 hyb capture kit in various plant genetics applications.
Antonelli, A., Clarkson, J.J., Kainulainen, K., Maurin, O., Brewer, G.E., Davis, A.P., … Baker, W.J. (2021). Settling a family feud: a high-level phylogenomic framework for the Gentianales based on 353 nuclear genes and partial plastomes. American Journal of Botany 108, 1143–1165.
Baker, W.J., Bailey, P., Barber, V., Barker, A., Bellot, S., Bishop, D., … Forest, F. (2021). A Comprehensive Phylogenomic Platform for Exploring the Angiosperm Tree of Life. Systematic Biology.
Brewer, G.E., Clarkson, J.J., Maurin, O., Zuntini, A.R., Barber, V., Bellot, S., … Baker, W.J. (2019). Factors Affecting Targeted Sequencing of 353 Nuclear Genes From Herbarium Specimens Spanning the Diversity of Angiosperms. Front. Plant Sci. 0.
Buerki, S., Callmander, M.W., Acevedo-Rodriguez, P., Lowry, P.P., Munzinger, J., Bailey, P., … Forest, F. (2021). An updated infra-familial classification of Sapindaceae based on targeted enrichment data. American Journal of Botany 108, 1234–1251.
Clarkson, J.J., Zuntini, A.R., Maurin, O., Downie, S.R., Plunkett, G.M., Nicolas, A.N., … Baker, W.J., (2021). A higher-level nuclear phylogenomic study of the carrot family (Apiaceae). American Journal of Botany 108, 1252–1269.
Gaynor, M.L., Fu, C.-N., Gao, L.-M., Lu, L.-M., Soltis, D.E., Soltis, P.S. (2020). Biogeography and ecological niche evolution in Diapensiaceae inferred from phylogenetic analysis. Journal of Systematics and Evolution 58, 646–662.
Hendriks, K.P., Mandáková, T., Hay, N.M., Ly, E., Huysduynen, A.H. van, Tamrakar, R., … Bailey, C.D. (2021). The best of both worlds: Combining lineage-specific and universal bait sets in target-enrichment hybridization reactions. Applications in Plant Sciences 9.
Johnson, M., Pokorny, L., Dodsworth, S., Botigue, L.R., Cowan, R.S., Devault, A., … Wickett, N. (2019). A Universal Probe Set for Targeted Sequencing of 353 Nuclear Genes from Any Flowering Plant Designed Using k-medoids Clustering. Systematic Biology 68(4): 594-606.
Larridon, I., Villaverde, T., Zuntini, A.R., Pokorny, L., Brewer, G.E., Epitawalage, N., … Baker, W.J. (2020). Tackling Rapid Radiations With Targeted Sequencing. Frontiers in Plant Science 10: 1.
Larridon, I., Zuntini, A.R., Barrett, R.L., Wilson, K.L., Bruhl, J.J., Goetghebeur, P., … Roalson, E.H. (2021). Resolving generic limits in Cyperaceae tribe Abildgaardieae using targeted sequencing. Botanical Journal of the Linnean Society 196, 163–187.
Larridon, I., Zuntini, A.R., Léveillé-Bourret, É., Barrett, R.L., Starr, J.R., Muasya, A.M., … Baker, W.J. (2021). A new classification of Cyperaceae (Poales) supported by phylogenomic data. Journal of Systematics and Evolution 59, 852–895.
Lee, A.K., Gilman, I.S., Srivastav, M., Lerner, A.D., Donoghue, M.J., Clement, W.L. (2021). Reconstructing Dipsacales phylogeny using Angiosperms353: issues and insights. American Journal of Botany 108, 1122–1142.
Maurin, O., Anest, A., Bellot, S., Biffin, E., Brewer, G., Charles-Dominique, T., … Lucas, E. (2021). A nuclear phylogenomic study of the angiosperm order Myrtales, exploring the potential and limitations of the universal Angiosperms353 probe set. American Journal of Botany 108(7): 1087–1111.
Murphy, B., Forest, F., Barraclough, T., Rosindell, J., Bellot, S., Cowan, R., … Cheek, M. (2020). A phylogenomic analysis of Nepenthes (Nepenthaceae). Molecular Phylogenetics and Evolution 144, 106668.
Ogutcen, E., Christe, C., Nishii, K., Salamin, N., Möller, M., Perret, M. (2021). Phylogenomics of Gesneriaceae using targeted capture of nuclear genes. Molecular Phylogenetics and Evolution 157, 107068.
Ottenlips, M.V., Mansfield, D.H., Buerki, S., Feist, M.A.E., Downie, S.R., Dodsworth, … Smith, J.F. (2021). Resolving species boundaries in a recent radiation with the Angiosperms353 probe set: the Lomatium packardiae/L. anomalum clade of the L. triternatum (Apiaceae) complex. American Journal of Botany 108, 1217–1233.
Pérez-Escobar, O.A., Dodsworth, S., Bogarín, D., Bellot, S., Balbuena, J.A., Schley, R.J., … Baker, W.J. (2021). Hundreds of nuclear and plastid loci yield novel insights into orchid relationships. American Journal of Botany 108, 1166–1180.
Pillon, Y., Hopkins, H.C.F., Maurin, O., Epitawalage, N., Bradford, J., Rogers, Z.S., … Forest, F. (2021). Phylogenomics and biogeography of Cunoniaceae (Oxalidales) with complete generic sampling and taxonomic realignments. American Journal of Botany 108, 1181–1200.
Shah, T., Schneider, J.V., Zizka, G., Maurin, O., Baker, W., Forest, F., … Larridon, I., (2021). Joining forces in Ochnaceae phylogenomics: a tale of two targeted sequencing probe kits. American Journal of Botany 108, 1201–1216.
Shee, Z. Q., D. G. Frodin, R. Cámara-Leret, L. Pokorny. (2020). Reconstructing the Complex Evolutionary History of the Papuasian Schefflera Radiation Through Herbariomics. Frontiers in Plant Science 11: 258.
Siniscalchi, C.M., Hidalgo, O., Palazzesi, L., Pellicer, J., Pokorny, L., Maurin, O., … Mandel, J.R. (2021). Lineage-specific vs. universal: A comparison of the Compositae1061 and Angiosperms353 enrichment panels in the sunflower family. Applications in Plant Sciences 9.
Starr, J.R., Jiménez-Mejías, P., Zuntini, A.R., Léveillé-Bourret, É., Semmouri, I., Muasya, M., … Larridon, I. (2021). Targeted sequencing supports morphology and embryo features in resolving the classification of Cyperaceae tribe Fuireneae s.l. Journal of Systematics and Evolution 59, 809–832.
Thomas, A.E., Igea, J., Meudt, H.M., Albach, D.C., Lee, W.G., Tanentzap, A.J. (2021). Using target sequence capture to improve the phylogenetic resolution of a rapid radiation in New Zealand Veronica. American Journal of Botany 108, 1289–1306.
Thomas, S.K., Liu, X., Du, Z.-Y., Dong, Y., Cummings, A., Pokorny, L., … Leebens-Mack, J.H. (2021). Comprehending Cornales: phylogenetic reconstruction of the order using the Angiosperms353 probe set. American Journal of Botany 108, 1112–1121.
Ufimov, R., Zeisek, V., Píšová, S., Baker, W.J., Fér, T., Loo, M., … Schmickl, R. (2021). Relative performance of customized and universal probe sets in target enrichment: A case study in subtribe Malinae. Applications in Plant Sciences 9, e11442.
Van Andel, T., Veltman, M. A., Bertin, A., Maat, H., Polime, T., Hille Ris Lambers, D., … Manzanilla, V.. (2019). Hidden Rice Diversity in the Guianas. Frontiers in Plant Science 10: 1161.
Wenzell, K.E., McDonnell, A.J., Wickett, N.J., Fant, J.B., Skogen, K.A. (2021). Incomplete reproductive isolation and low genetic differentiation despite floral divergence across varying geographic scales in Castilleja. American Journal of Botany 108, 1270–1288.
Zuntini, A.R., Frankel, L.P., Pokorny, L., Forest, F., Baker, W.J. (2021). A comprehensive phylogenomic study of the monocot order Commelinales, with a new classification of Commelinaceae. American Journal of Botany 108, 1066–1086.








